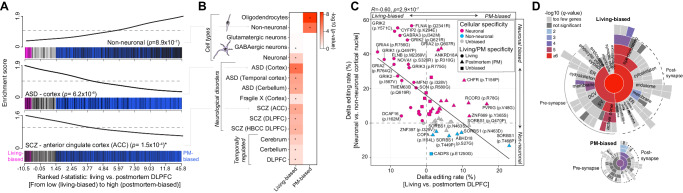
Fig. 4. Annotating dynamically regulated sites between living and postmortem DLPFC.
A CAMERA enrichment scores (y axes) for three candidate sets of A-to-I editing sites along a ranked list of differentially edited sites (t statistics; x axis) between living and postmortem (PM) DLPFC, from highly living-biased (right; pink) to highly PM-biased (left; blue) (x axes). Enrichment plots for non-neuronal sites (top, PM biased), sites disrupted in autism spectrum disorder (ASD) cortex (middle, living biased), and those disrupted in schizophrenia (SCZ) ACC (bottom, living biased). B Summary of all multiple test-corrected p values (−log10) for all sets of RNA editing sites across cell types, neurodevelopmental disorders, and brain development. A, B CAMERA gene set enrichment p values, quantifying the statistical significance of overrepresented A-to-I sites within the ranked living versus postmortem data. C Pearson’s correlation and scatterplot of delta editing rates for cell-specific recoding sites (y axis) versus delta editing rates for living/PM differences (x axis). Y axis description: Fluorescence activated nuclei sorted (FANS) neurons and non-neuronal cell populations were collected from 10 postmortem donors across five cortical regions (see Supplemental Note 1). D SynGO synaptic enrichment (−log10 q value) for genes harboring living-biased editing sites (top) and genes harboring postmortem-biased sites (bottom). RNA-seq analysis encompassed 164 and 233 biologically independent samples from living and postmortem sources, respectively.