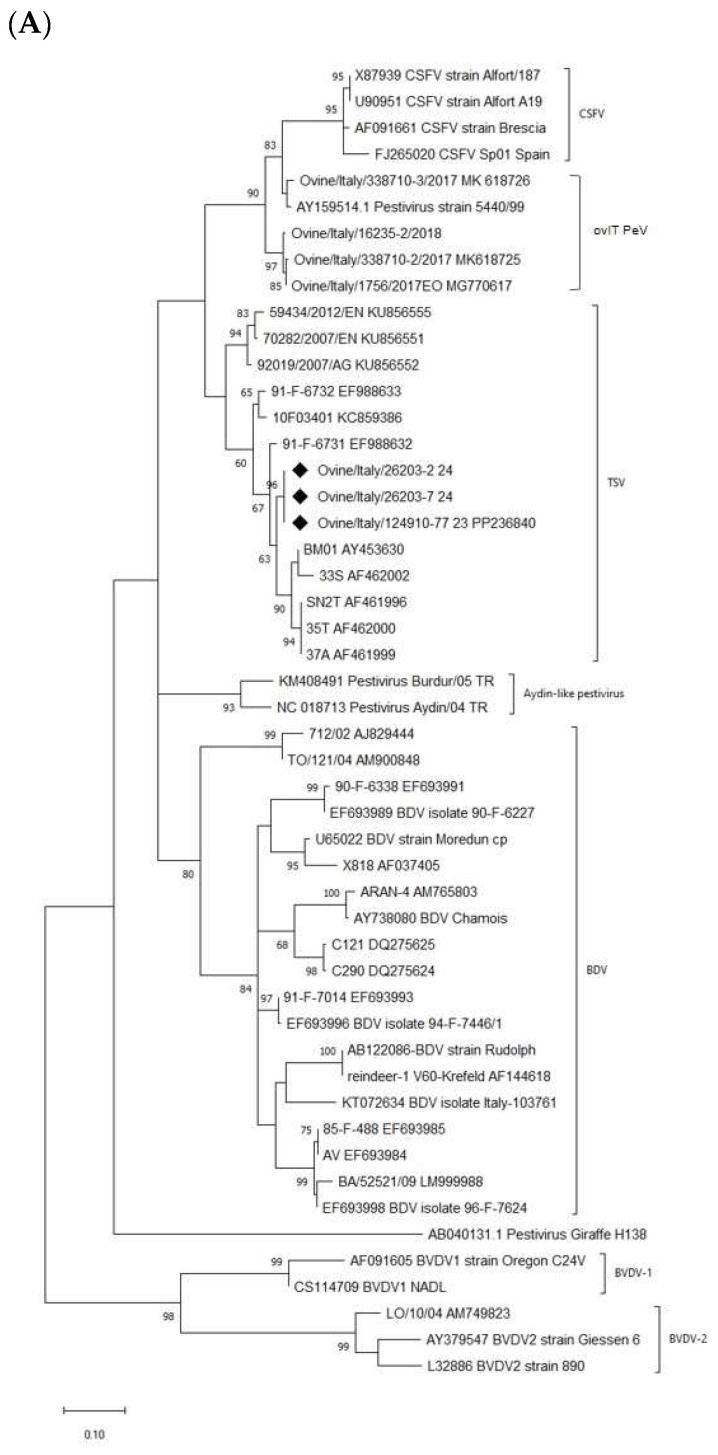
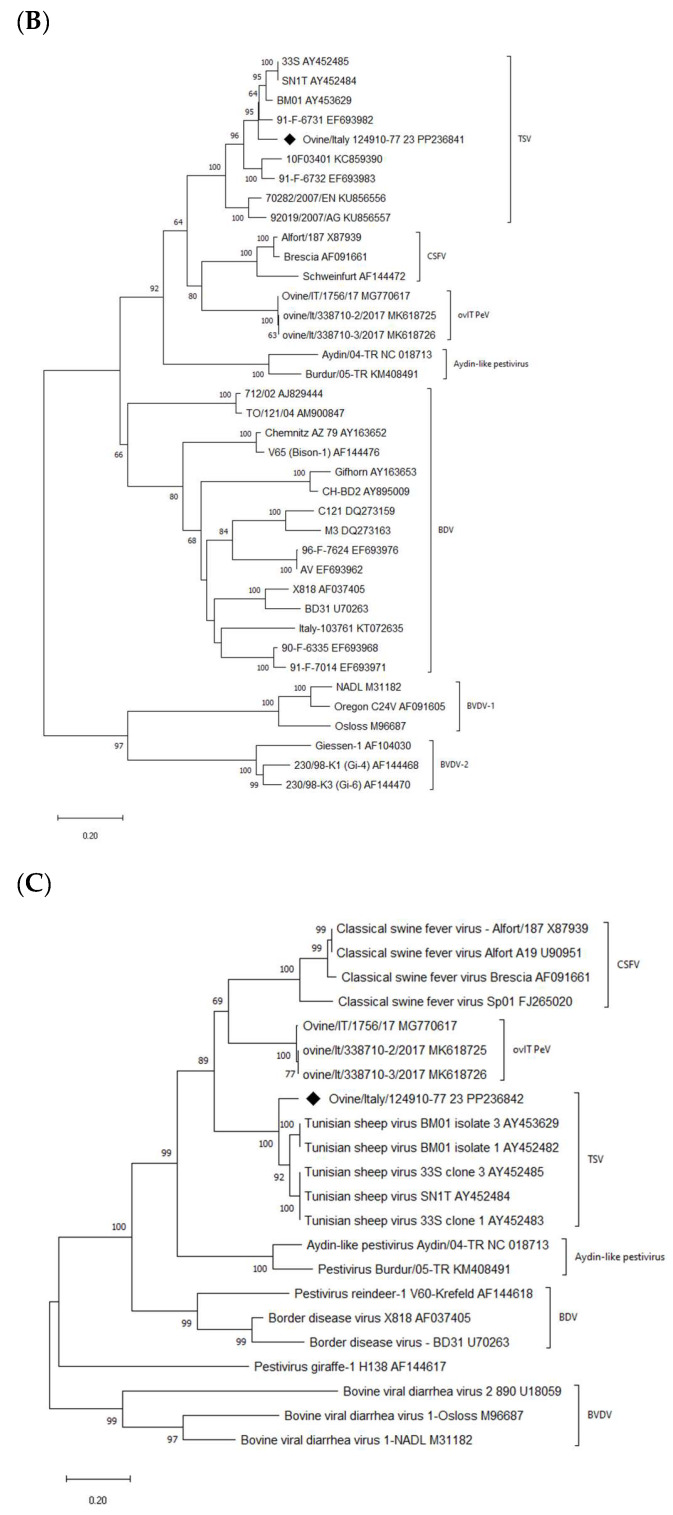
Figure 1.
The phylogenetic trees are based on (A) the partial 5′-UTR, (B) partial Npro, and (C) complete E2 regions sequencing of the TSV strain detected in the survey and the TSV isolates and other pestivirus sequences deposited in GenBank. Molecular analyses were performed with MEGA v.X software with bootstrap analysis (1000 replicates) using the Maximum Likelihood method based on the Kimura 2 parameter (G), Tamura-Nei (G + I), and General Time Reversible (G + I) models, respectively, for 5′-UTR, Npro, and E2. Bootstrap values > 60% are shown. Sequences generated in this study are indicated with a black rhombus. Published sequences are identified by strain and GenBank accession number.