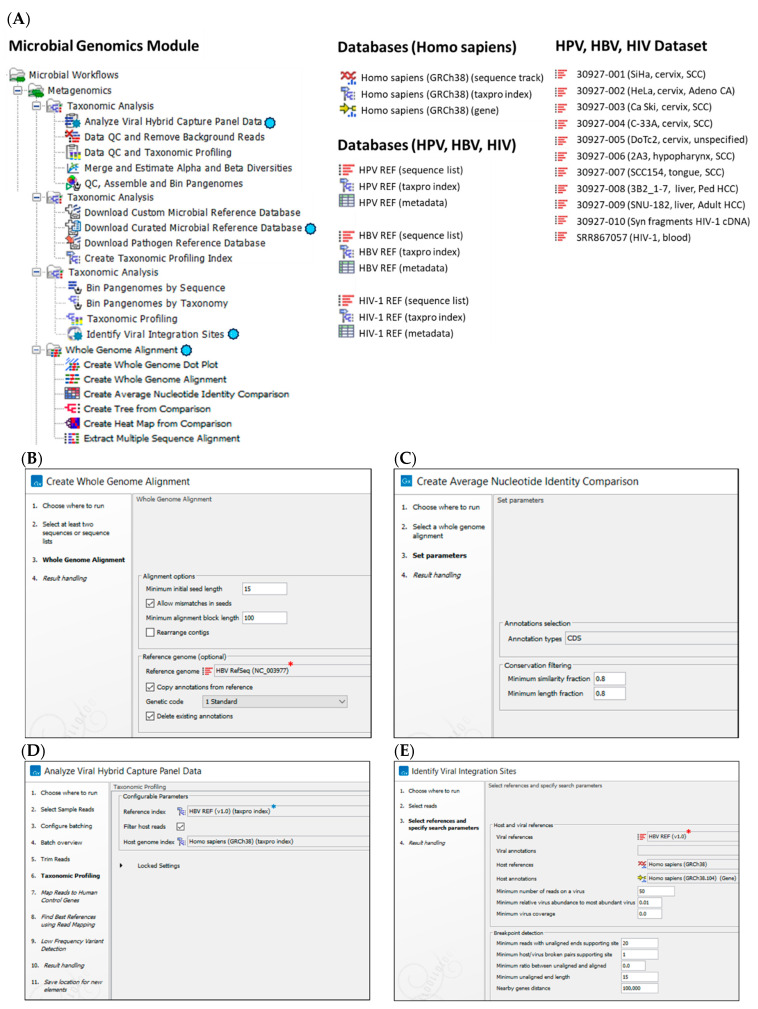
Figure 2.
Bioinformatics methods. (A) CLC Microbial Genomics Module, databases and dataset used for Whole-Genome Alignment (WGA), Viral Hybrid Capture (VHC) data analysis, and Viral Integration Site (VIS) analysis. Primary workflows and tools used for this study are designated by the virus icon ( ); (B) WGA workflow steps (1–4) with user-defined parameter settings for WGA and annotation, e.g., HBV RefSeq (*) genome; (C) Create Average Nucleotide Identity Comparison workflow inputs the WGA file for quantification of the similarity between genomes, and outputs a pairwise comparison matrix; (D) VHC workflow steps (1–11) with user-defined parameter settings for Taxonomic Profiling (*), e.g., HBV reference index and host genome index; (E) VIS workflow steps (1–4) with selected HBV (*) and host reference genome databases and user-defined search parameters entered for this study.
); (B) WGA workflow steps (1–4) with user-defined parameter settings for WGA and annotation, e.g., HBV RefSeq (*) genome; (C) Create Average Nucleotide Identity Comparison workflow inputs the WGA file for quantification of the similarity between genomes, and outputs a pairwise comparison matrix; (D) VHC workflow steps (1–11) with user-defined parameter settings for Taxonomic Profiling (*), e.g., HBV reference index and host genome index; (E) VIS workflow steps (1–4) with selected HBV (*) and host reference genome databases and user-defined search parameters entered for this study.