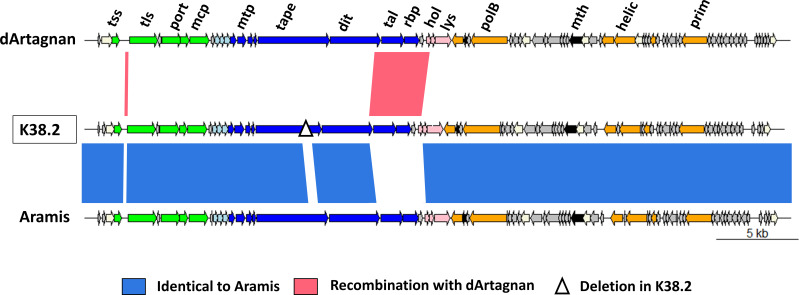
Fig 7.
Mutational events detected in K38.2, which was mainly derived from Enterococcus phage Aramis having recombined with the closely related Enterococcus phage dArtagnan. The three genomes were represented using genoplotR (46). Gene functions are color coded (and abbreviated): orange, nucleic acid metabolism (polB, B family polymerase; helic, helicase; prim, primase); green, DNA packaging and head (tss and tls, terminase small and large subunits; port, portal; mcp, major capsid protein); dark blue, tail [mtp, major tail protein, tape, tail length tape measure protein; dit, distal tail protein; tal, tail-associated lysozyme protein, rbp, (putative) receptor-binding protein]; light blue, connector; pink, lysis (lys, endolysin; hol, holin); white, HNH endonuclease; black, auxiliary metabolism (mth, metallo-hydrolase); gray, hypothetical proteins. The mutational events are indicated below.