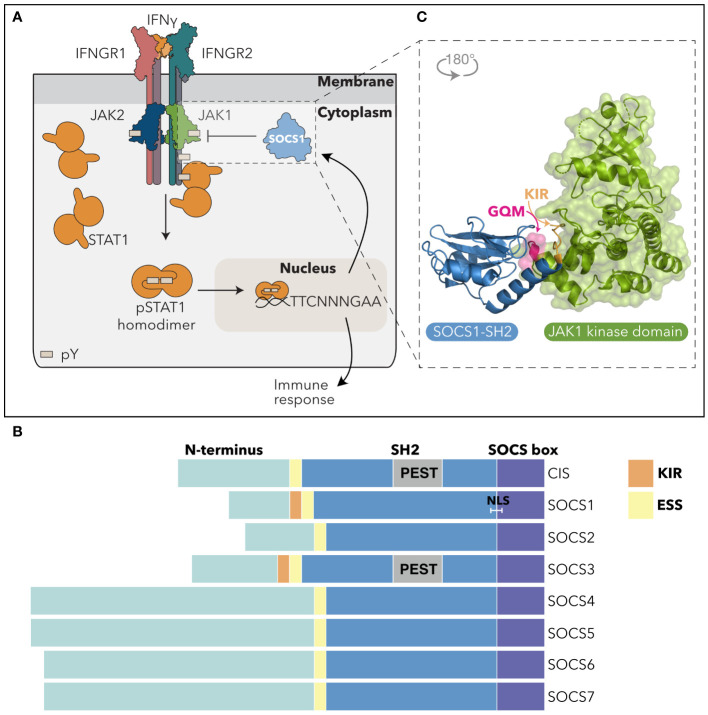
Figure 1.
SOCS1 inhibits the IFNγ-driven JAK-STAT pathway. (A) SOCS1 negatively regulates IFNγ signaling. Upon engagement with the IFNGR complex, IFNγ induces tyrosine-phosphorylation and activation of JAK1 and JAK2. Activated JAK1/2 phosphorylate intracellular Tyr motifs in the IFNGR1, leading to STAT1 recruitment via the STAT1-SH2 domain and its subsequent phosphorylation by JAK. pSTAT1 dimers undergo a conformation change and translocate to the nucleus, binding to gamma interferon activation sites (GAS) to regulate transcription and drive a cellular response. SOCS1 is an IFNγ-response gene and is induced to inhibit IFNγ signaling in a classic negative feedback loop. pY = phosphotyrosine; TTCNNNGAA = IFNγ-activated promoter sequences. (B) SOCS domain architecture. The SOCS domain architecture consists of an unstructured N-terminal region of variable length (teal), a central SH2 domain (blue) and a C-terminal SOCS box motif (purple). SOCS1 and SOCS3 are distinguished by a KIR that precedes the ESS and SH2 domain region. The CIS and SOCS3 SH2 domains contain a PEST insertion (grey), while SOCS1 contains a putative NLS. (C) SOCS1 inhibition of JAK. Non-canonical binding of the SOCS1-SH2 domain (blue) to the JAK-GQM motif (pink), enables blocking of JAK-enzymatic activity via the SOCS1-KIR (orange). PDB: 6C7Y (5). SH2, Src-Homology 2 domain; ESS, Extended SH2 Sequence; KIR, Kinase Inhibitory Region; PEST, sequence rich in proline (P), glutamic acid (E), serine (S), and threonine (T); NLS*, nuclear localisation signal.