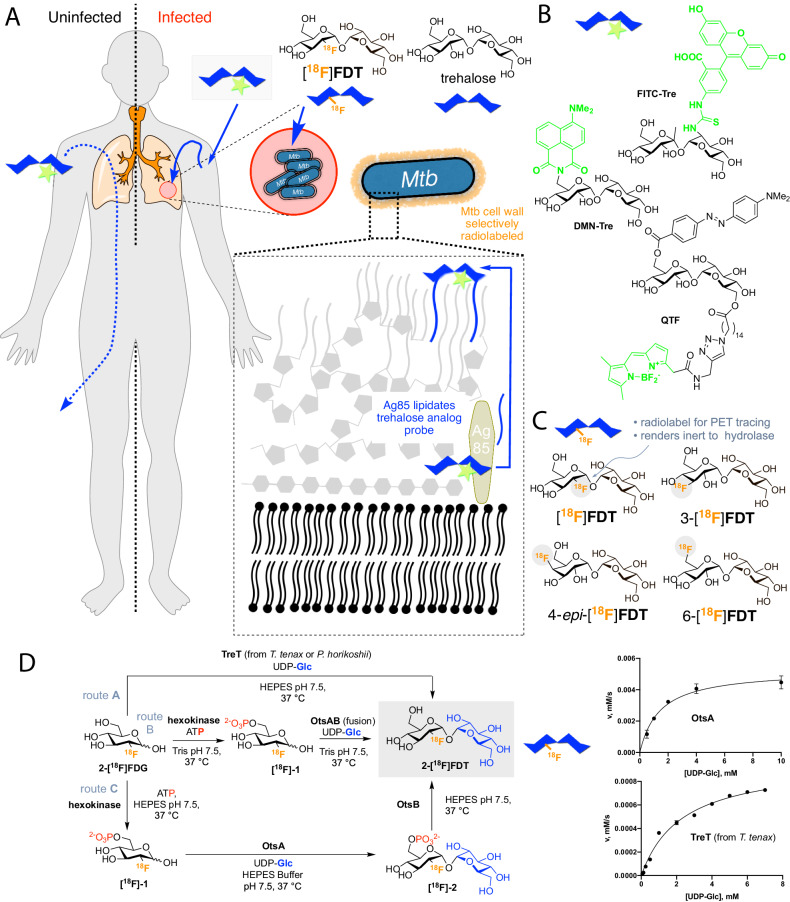
Fig. 1. A Strategy for Non-Invasive Imaging Reporters using Trehalose-based, TB-specific Probes Derived Directly from [18F]FDG.
A Trehalose (blue) in Mtb is found in the outer portion of the mycobacterial cell envelope as its corresponding mycolate glycolipids. The biosynthesis of trehalose mycolates (lipidation) is catalyzed specifically in Mtb by abundant membrane-associated Antigen 85 (Ag85a, Ag85b and Ag85c) enzymes. The lack of naturally occurring trehalose in mammalian hosts as well as the uptake of exogenous trehalose by Mtb suggests that it could function as both a highly specific and sensitive probe, allowing here the development of an in vivo TB-specific, PET-radiotracer analog [18F]FDT that selectively labels lesions (red) in infected organisms (right). Uninfected organisms (left) do not process trehalose and so probe is not retained. B Prior work has established that Ag85s are sufficiently plastic in their substrate scope that they can process, for example, fluorescent analogs of trehalose, allowing them to be metabolically incorporated into the mycobacterial outer membrane in vitro for labeling. Fluorescence-based methods are not yet amenable to effective, non-invasive imaging in vivo. C Four 18F-labeled variants of trehalose were tested in which each of the available hydroxyls (OH−2, 3, 4, 6) were converted in turn to 18F. D Enzymatic synthesis of [18F]FDT. Three parallel routes (A, B and C) were evaluated for efficiency, rate and yield. Route A: TreT-mediated synthesis was evaluated using enzymes from two different sources (from T. tenax or P. horikoshii). Both proved functional but gave lower turnover under a range of conditions (see pseudo-single substrate plot for TreT (from T. tenax), lower right – See Fig. S1 and Source Data File for further details of kinetics). [Reagents and Conditions: 50 mM HEPES, 100 mM NaCl and 10 mM MgCl2, pH 7.5]. Route B: OtsAB-fusion enzyme-mediated synthesis was explored. An OtsAB fusion protein was constructed but proved to be more difficult to express and less stable under typical reaction conditions [Reagents and Conditions: 50 mM HEPES, 100 mM NaCl and 10 mM MgCl2, pH 7.5 C] Route C: Although a three-step, three-enzyme route, Route C proved to be more flexible and reliable. The selectivity of the biocatalysts allowed this to be performed in a convenient one-pot manner. The greater stability of enzyme components and the ability to vary catalyst amounts to control flux led to its choice over Route B. The higher turnovers and efficiencies led to its choice over Route A (see pseudo-single substrate plot for OtsA, upper right – See Fig. S1 and Source Data File for further details of kinetics). [Reagents and Conditions: 50 mM HEPES, 100 mM NaCl and 10 mM MgCl2, pH 7.5.]. The data points are average values from three replicates with error bars ± SD(n = 3).