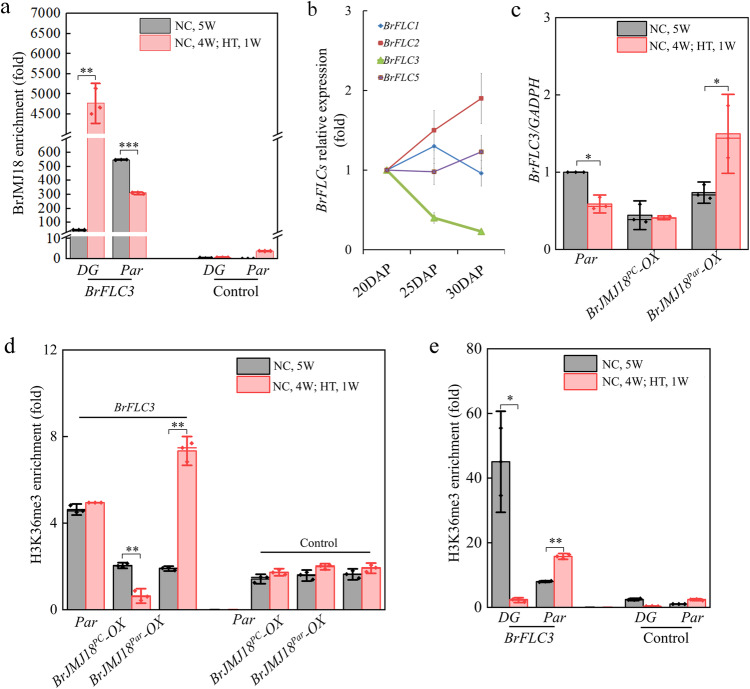
Fig. 5. Overexpression of BrJMJ18Par moderates flowering by altering the expression of BrFLC3.
a Chromatin immunoprecipitation (ChIP) analysis of the BrJMJ18 level across BrFLC3 in BrJMJ18Par-carrying Par and BrJMJ18PC-carrying DG plants. The 5th−7th young and healthy rosette leaves of five-week-old plants were used for the analysis. Rabbit IgG was used as a control. GADPH was used as a BrJMJ18-independent control. Control is a locus gene desert region where BrJMJ18 does not bind. The values are the mean ± standard deviation from three biological replicates. Asterisks indicate significant differences between NC and HT, two-tailed Student’s t-test (**0.01 < p < 0.05, ***p < 0.001). DG, p = 0.0016; Par, p = 0.00027. b Expression patterns of BrFLC1, 2, 3 and 5 in Par before and after bolting under natural field conditions. The 5th−7th young and healthy rosette leaves were used for qPCR test. The values are the mean ± standard deviation from three biological replicates. The Par bolted at 25DAP. DAP, days after planting (14-day Par seedlings grown in cultivation pots in greenhouse were transplanted into natural field). c BrFLC3 expression patterns in BrJMJ18PC/Par- overexpressing plants. The 5th−7th young and healthy rosette leaves of Par plants were used for qPCR test. The values are the mean ± standard deviation from three biological replicates. Asterisks indicate significant differences between NC and HT, two-tailed Student’s t test (*0.01 < p < 0.05). BrJMJ18PC-OX, p = 0.0032; BrJMJ18Par-OX, p = 0.0023. d ChIP analysis of H3K36me3 enrichment on the BrFLC3 locus in BrJMJ18 overexpression Par lines. The values are the mean ± standard deviation from three biological replicates. Asterisks indicate significant differences between NC and HT, two-tailed Student’s t test (** 0.001 < p < 0.01). Par, p = 0.012; BrJMJ18Par-OX, p = 0.034. e The H3K36me3 level at BrFLC3 in DG (BrJMJ18PC-carrying) and Par (BrJMJ18Par-carrying). The values are the mean ± standard deviation from three biological replicates. Asterisks indicate significant differences between NC and HT, two-tailed Student’s t-test (*p < 0.05, **0.001 <p < 0.01,). DG, p = 0.021; Par, p = 0.0016. Source data are provided as a Source Data file.