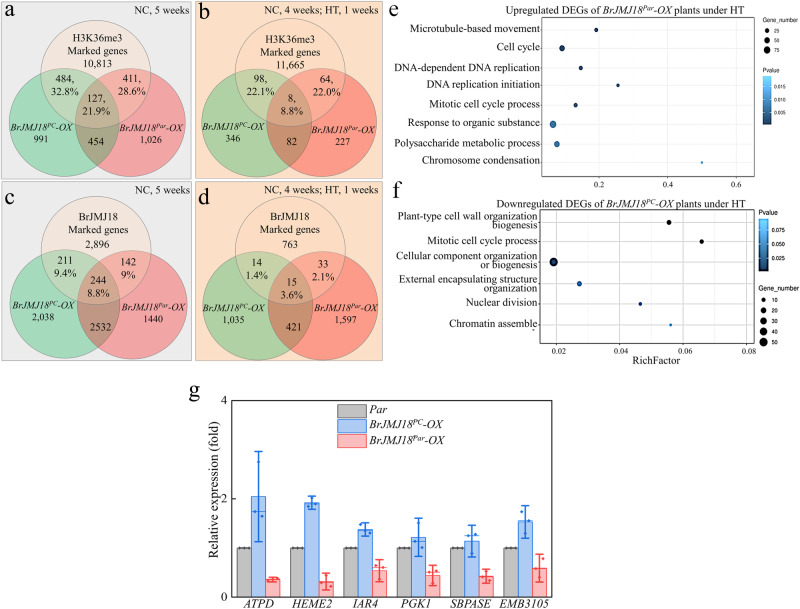
Fig. 7. Integrated analysis of RNA-seq and ChIP-seq data reveled BrJMJ18Par modulates chlorophyll biosynthesis r under high temperature.
a Diagram showing the overlap of genes targeted by BrJMJ18PC and BrJMJ18Par, respectively, and reported H3K36me3-regulated genes under NC. b Diagram showing the overlap of genes targeted by BrJMJ18PC and BrJMJ18Par, respectively, and reported H3K36me3-regulated genes under HT. c Diagram showing the overlap of the DEGs of BrJMJ18PC-OX and BrJMJ18Par-OX plants, respectively, and the identified BrJMJ18-targeted genes under NC. Genes showing a 2-fold change within a 95% confidence interval were considered to be differentially expressed. d Diagram showing the overlap of the DEGs of BrJMJ18PC-OX and BrJMJ18Par-OX plants, respectively, and the identified BrJMJ18-targeted genes under HT. Genes showing a 2-fold change within a 95% confidence interval were considered to be differentially expressed. e, f Functional categorization by Gene Ontology of the DEGs of BrJMJ18PC-OX and BrJMJ18Par-OX plants under HT. Only significantly enriched entries are shown. e, GO analysis demonstrated that the upregulated DEGs in BrJMJ18Par-OX plants under HT were mainly enriched in cell division. f, the cell division-related GO entries were enriched in the downregulated DEGs in the BrJMJ18PC-OX line. g Expression of heat stress-related genes reported by Zhang et al. in Par and BrJMJ18-OX plants. 5-week-old plants grown under NC, and 4-week-old plants grown under NC followed by 1 week under HT were used for Q-PCR. Relative to Par plants, all six genes exhibited slight induction in BrJMJ18PC-OX plants under high temperatures, while they were significantly downregulated in BrJMJ18Par-OX plants. Photosynthesis-related gene BraA9g029800.3 C (ATP synthase DELTA-subunit gene, ATPD), porphyrin and chlorophyll metabolism-related gene BraA04g028660.3 C (Uroprophyrinogen decarboxylase, HEME2) and carbon metabolism-related genes BraA07g034950.3 C (Pyruvate dehydrogenase E1a-like subunit, IAR4), BraA03g035490.3 C (Phosphoglycerate kinase 1, PGK1), BraA07g0022160.3 C (Sedoheptulose- bisphosphatase, SBPASE), BraA08g004460.3 C (Embryo defective 3105, EMB3105). GADPH was used as the internal control. The values are the mean ± standard deviation from three biological replicates. Source data are provided as a Source Data file.