Abstract
Mycotoxins, which are secondary metabolites produced by toxicogenic fungi, are natural food toxins that cause acute and chronic adverse reactions in humans and animals. The genus Fusarium is one of three major genera of mycotoxin‐producing fungi. Trichothecenes, fumonisins, and zearalenone are the major Fusarium mycotoxins that occur worldwide. Fusarium mycotoxins have the potential to infiltrate the human food chain via contamination during crop production and food processing, eventually threatening human health. The occurrence and development of Fusarium mycotoxin contamination will change with climate change, especially with variations in temperature, precipitation, and carbon dioxide concentration. To address these challenges, researchers have built a series of effective models to forecast the occurrence of Fusarium mycotoxins and provide guidance for crop production. Fusarium mycotoxins frequently exist in food products at extremely low levels, thus necessitating the development of highly sensitive and reliable detection techniques. Numerous successful detection methods have been developed to meet the requirements of various situations, and an increasing number of methods are moving toward high‐throughput features. Although Fusarium mycotoxins cannot be completely eliminated, numerous agronomic, chemical, physical, and biological methods can lower Fusarium mycotoxin contamination to safe levels during the preharvest and postharvest stages. These theoretical innovations and technological advances have the potential to facilitate the development of comprehensive strategies for effectively managing Fusarium mycotoxin contamination in the future.
Keywords: climate change, Fusarium mycotoxin, management strategy, mycotoxin detection
INTRODUCTION
Fungi are a kind of eukaryote with a vegetative structure that can be filamentous or unicellular. Their cell walls are composed of chitin, chitosan, or polysaccharides, and they reproduce by spores that are produced asexually or sexually. Fungi are ubiquitous in the biosphere and the second most species‐rich eukaryotic organisms after insects 1 . They play a pivotal role in the intricate processes of the nutrient cycle and ecosystem balance and are consumed as food and used in the production of industrial materials 2 . Similar to viruses and bacteria, fungi also have negative effects on humans. More than 500 fungal species are capable of infecting the human body, which causes at least 1.5 million deaths globally each year 3 . In plantation crop production, plant pathogenic fungi can cause economic losses of upward of hundreds of billions of dollars each year. Plant pathogenic fungi adversely affect crop growth, yield, and quality, and mycotoxin contamination is one of the most important causes of quality degradation. 4
Mycotoxins are natural toxic compounds with low molecular weight (often less than 1000 Da) that are synthesized by fungi and have the characteristics of nephrotoxicity, hepatotoxicity, carcinogenicity, teratogenicity, immune toxicity, neurotoxicity, genotoxicity, mutagenicity, cytotoxicity, reproductive toxicity, alimentary canal toxicity, dermal toxicity, and so on 5 , 6 . Mycotoxin exposure, whether in humans or in animals, even at low concentrations, can lead to acute or chronic diseases and, in certain instances, death. Cereals, nuts, fruits, spices, legumes, and their products or byproducts are most likely to contain mycotoxins. Based on a previous estimation by the Food and Agriculture Organization (FAO) of the United Nations, approximately 25% of cereals produced worldwide are contaminated by mycotoxins 7 . However, a recent study conducted by Eskola et al. revealed that the contamination rate could be 60%–80% 8 . Mycotoxins enter the food chain via fungal infections of crops, which can occur at any phase of crop production, including planting, harvesting, and storage. Mycotoxins can be introduced into the human body directly via the ingestion of contaminated foods or derived products, or indirectly through the consumption of eggs, edible offal, meat, milk, and related products from livestock that consume contaminated feed 8 .
Mycotoxins are not essential to the growth or development of fungi, but they seem to be a way for fungi to reduce the number of superfluous precursors 9 . It has also been suggested that mycotoxins contribute to the defensive tactics of mycotoxigenic fungi against other microorganisms. Moreover, mycotoxins exert a substantial influence on the pathogenicity, aggressiveness, and virulence of mycotoxigenic fungi 10 . More than 400 varieties of mycotoxins have been identified, but aflatoxin (AF), ochratoxins, fumonisins (FUMs), trichothecenes (TRIs), patulin, citrinin (CIT), and zearalenone (ZEA) are those most inextricably linked to agriculture, economics, and public health 5 , 6 . There are three dominant toxigenic fungal genera: Aspergillus, Fusarium, and Penicillium 6 . Fusarium species usually infect crops and produce mycotoxins before or immediately after harvest, while the Aspergillus and Penicillium species are more commonly associated with foods during drying and storage 5 .
Fusarium, a prominent genus of plant pathogenic and mycotoxin‐producing fungi worldwide, targets various plant parts, including grains, seedlings, heads, roots, and stems (Figure 1), causing yield loss and quality reduction of crops 11 . For example, Fusarium head blight (FHB) is a highly destructive disease that affects cereals and results in both significant yield reduction and a negative influence on grain quality 12 . Currently, Fusarium comprises more than 300 phylogenetically distinct species and has been classified into 23 informal species complexes 13 . Different Fusarium species have various abilities to produce mycotoxins. The Fusarium sambucinum species complex includes many devastating plant pathogens, most of which are important mycotoxin producers (Figure 2); the three predominant classes of mycotoxins synthesized by Fusarium species are FUMs, TRIs, and ZEA. Moreover, a type of TRI analog, deoxynivalenol (DON), is consistently regarded as a significant concern in the realm of food safety due to its prevalent occurrence as a grain contaminant. Due to a lack of adequate understanding, other toxic Fusarium metabolites, which are commonly referred to as emerging mycotoxins and include beauvericin (BEA), enniatins, fusaproliferin, fusaric acid, fusarins, and moniliformin (MON), neither undergo routine determination nor are subject to legislative regulation 11 . Furthermore, Fusarium mycotoxins can be converted into modified forms, known as modified mycotoxins, by plants, microorganisms, and chemical or physical approaches during processing, such as acid, alkali, heat, pressure, and irradiation. The modified mycotoxins can coexist with their parent forms 14 . Among these, the modified mycotoxins generated as a result of plant defense mechanisms, primarily via glucosylation catalyzed by uridine diphosphate‐glucosyltransferases, are known as masked mycotoxins 15 . The glucose conjugates of mycotoxins, the most commonly identified masked mycotoxins in current research, include DON‐3‐glucoside (DON‐3G), T‐2‐toxin‐3‐glucoside, HT‐2‐toxin‐3‐glucoside, nivalenol‐3‐glucoside (NIV‐3G), ZEA‐14G, α‐zearalenol‐14‐glucoside (α‐ZEL‐14‐G), and β‐ZEL‐14‐G. Although the limited available information suggests that masked mycotoxins show lower toxicity than their parent forms, the free forms of masked mycotoxins can still cause unpredicted toxicity via hydrolysis by mammalian gut microorganisms 15 .
Figure 1.
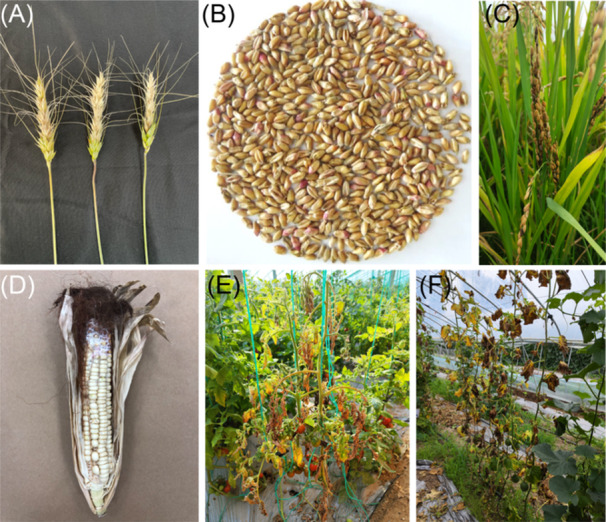
Examples of plant Fusarium diseases. (A) Fusarium head blight (FHB) of wheat caused by Fusarium graminearum (photo credit: Dr. Jie Wang). (B) Diseased wheat kernels due to FHB (photo credit: Dr. Binnian Tian). (C) Rice spikelet rot disease caused by Fusarium proliferatum (photo credit: Dr. Yanpo Yao). (D) Fusarium ear rot of corn caused by Fusarium verticillioides (photo credit: Dr. Zheng Qu). (E) Fusarium wilt of cherry tomato caused by Fusarium oxysporum (photo credit: Dr. Ying Zhao). (F) Fusarium wilt of melon caused by F. oxysporum (photo credit: Dr. Ying Zhao).
Figure 2.

The varieties of mycotoxin production in Fusarium species. On the left side, the cladogram depicts phylogenetic relationships among the chosen set of Fusarium species. On the right side, the circle represents the mycotoxin production abilities of Fusarium species. The cladogram was constructed by maximum likelihood analysis using the RPB1 + RPB2 dataset described by O'Donnell et al. 16 in RAxML 8.2.10. Support for branches was determined through bootstrap analysis (1000 replicates), and only values of more than 60 are shown. The distribution of mycotoxin production was based on the work of Munkvold et al. 11 . White space indicates that there are no chemical or genetic evidence for mycotoxin production.
In this review, we outline the main types of Fusarium mycotoxins and elucidate their impact on food contamination as well as associated hazards. We then review the interactions between climate change (CC) and Fusarium mycotoxin risks. Finally, we focus on the detection technologies and management strategies for Fusarium mycotoxin contamination. This review provides a reliable reference source for Fusarium mycotoxin control.
MAIN FUSARIUM MYCOTOXINS
TRIs
The most extensive and economically important cluster of Fusarium mycotoxins are the TRIs, which include over 200 compounds. TRIs are tetracyclic sesquiterpenoid substances characterized by a single six‐membered ring with a single oxygen atom flanked by two carbon rings (Figure 3A). This core structure shows a double bond between C‐9 and C‐10, along with an epoxide ring at the C‐12 and C‐13 positions 17 , 18 , 19 . According to the various patterns of oxygenation and esterification at the C‐3, C‐4, C‐7, C‐8, and C‐15 positions, TRIs can be classified into four groups: Types A, B, C, and D. Fusarium species produce Type A and Type B TRIs, which are concentrated in the F. sambucinum and Fusarium incarnatum‐equiseti species complexes, respectively 11 , 19 . Representative Type A TRIs include T‐2 toxin (T‐2), HT‐2 toxin (HT‐2), neosolaniol (NEO), diacetoxyscirpenol (DAS), monoacetoxyscirpenol (MAS), and NX‐2. Type B TRIs are represented by DON, NIV and their acetyl derivatives: 3‐acetyldeoxynivalenol (3ADON), 15‐acetyldeoxynivalenol (15ADON), and 4‐acetylnivalenol (4ANIV). The toxic effects of TRIs can be teratogenic, nephrotoxic, hepatotoxic, cytotoxic, alimentary canal toxic, genotoxic, and immune toxic 20 .
Figure 3.
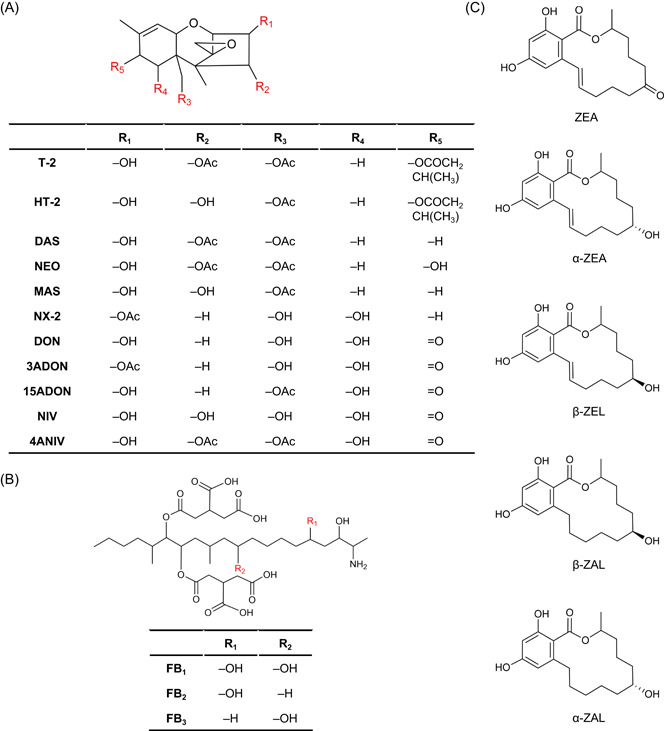
Chemical structures of primary Fusarium mycotoxins. (A) Chemical structures of Types A and B trichothecenes produced by Fusarium, such as diacetoxyscirpenol (DAS), deoxynivalenol (DON), HT‐2 toxin (HT‐2), T‐2 toxin (T‐2), monoacetoxyscirpenol (MAS), neosolaniol (NEO), and nivalenol (NIV), the acetylated derivatives of DON and NIV (3ADON, 15ADON, 4ANIV), and the new trichothecene mycotoxin NX‐2. (B) Chemical structures of B‐series fumonisins, which show structural variations based on the presence or absence of hydroxyl groups at C‐5 (R1) and C‐10 (R2). (C) Chemical structure of zearalenone and its derivatives.
In Fusarium, TRI biosynthetic enzymes are encoded by a total of 15 TRI genes, which are distributed across three different loci on different chromosomes: the single‐gene TRI101 locus, the two‐gene TRI1‐TRI16 locus, and the 12‐gene core TRI cluster (Figure 4A) 21 , 22 . In F. graminearum, the TRI16 homolog is rendered nonfunctional as a result of multiple insertions and deletions within its coding region. Table 1 shows the phenotypes of mutants with TRI gene disruption or deletion in F. sporotrichioides. TRI5 catalyzes the cyclization of farnesyl pyrophosphate (FPP) into trichodiene (TDN), which represents the initial enzymatic step in TRI biosynthesis (Figure 4B). TDN is subsequently transformed into calonectrin (CAL) by TRI4, TRI101, TRI11, and TRI3. The catalytic reactions from FPP to CAL are conserved across Fusarium species that synthesize Type A and Type B TRIs. The allelic variants of TRI1 are responsible for the significant structural disparities between Type A and Type B TRIs. In F. sporotrichioides, TRI1 exclusively catalyzes hydroxylation at C‐8, resulting in Type A TRIs, while F. graminearum TRI1 mediates hydroxylation at both C‐7 and C‐8, resulting in Type B TRIs 17 , 21 , 22 .
Figure 4.
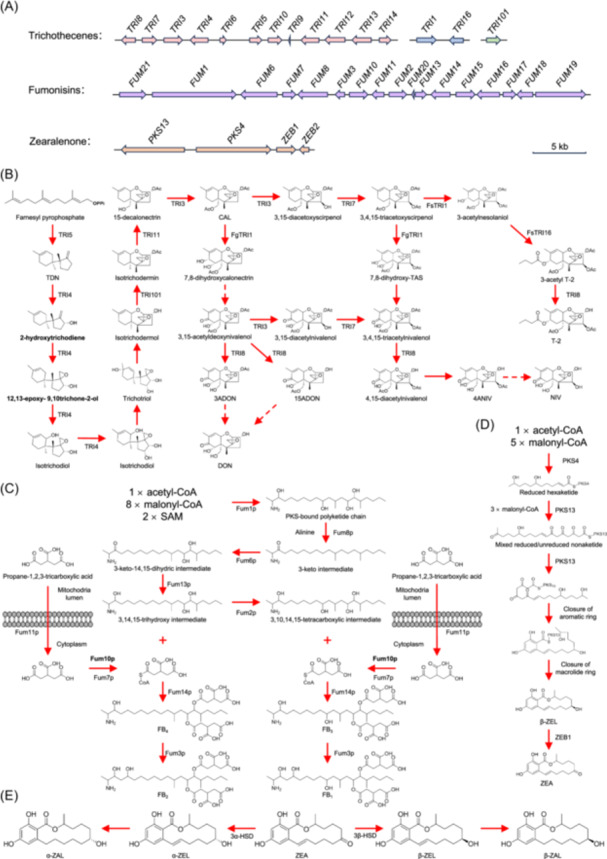
The formation approaches of Fusarium mycotoxins. (A) Mycotoxin biosynthetic genes and gene clusters in Fusarium. Arrows denote the location and transcriptional orientation of genes, while the corresponding gene name is provided adjacent to each arrow. (B) The proposed pathway of TRI biosynthesis. The 3,4,15‐triacetoxyscirpenol is an important branching point. One path leads to a Type A TRI, and the other leads to a Type B TRI. In F. sporotrichioides, FsRI1 exclusively catalyzes hydroxylation of 3,4,15‐triacetoxyscirpenol at C‐8, resulting in Type A TRIs, while in F. graminearum, FgTRI1 mediates hydroxylation of 3,4,15‐triacetoxyscirpenol at both C‐7 and C‐8, resulting in Type B TRIs. Dashed arrows denote steps where gene assignment has not been established. (C) The proposed pathway of FUM biosynthesis. Fum11p, a tricarboxylate transporter, transports tricarboxylic acid precursors out of the inner mitochondrial lumen for FUM biosynthesis, while Fum7p and Fum10p catalyze the conversion of tricarboxylic acid precursors into acetyl CoA‐activated tricarballylic acid. This product is then esterified to the polyketide backbone at C‐14 and C‐15 by Fum14p to produce either FB3 or FB4. Finally, the hydroxylation of FB3 and FB4 by Fum3p yields FB1 and FB2, respectively. (D) The proposed pathway of ZEA biosynthesis. The biosynthesis begins with PKS4, which facilitates the condensation of carbons derived from one acetyl‐CoA and five malonyl‐CoAs. PKS13 completes the polyketide backbone as an extender unit and is responsible for cyclization and aromatization. As the final step, β‐ZEL is converted into ZEA by ZEB1. (E) The formation pattern of metabolites of ZEA. The C6 keto group of ZEA is reduced to yield α‐ZEL and β‐ZEL, followed by a subsequent reduction at the C11–C12 double bond that results in the formation of α‐ZAL and β‐ZAL, respectively. Those proposed biosynthesis and metabolites pathways were derived from the works of McCormick et al. 17 , Alexander et al 21 , Nahle et al 23 and EFSA Panel on Contaminants in the Food Chain (CONTAM) et al 74 . CAL, calonectrin; FgTRI1, F. graminearum TRI1; FsTRI1, F. sporotrichioides TRI1; FUM, fumonisin; PKS, polyketide synthase; SAM, S‐adenosyl methionine; TRI, trichothecenes; ZEA, zearalenone.
Table 1.
Function of Fusarium mycotoxin biosynthesis genes and phenotypes of their mutants.
| Mycotoxin | Gene | Predicted function | Mutant phenotype |
|---|---|---|---|
| TRIs | TRI8 | TRI‐3‐O‐esterase | 3‐acetyl T‐2, TAS |
| TRI7 | TRI‐4‐O‐acetyltransferase | HT‐2 | |
| TRI3 | TRI‐15‐O‐acetyltransferase | 15‐decalonectrin, 3,15‐didecalonectrin | |
| TRI4 | Trichodiene oxygenase | Trichodiene | |
| TRI6 | Transcription factor | Low levels of trichodiene | |
| TRI5 | Trichodiene synthase | No TRIs | |
| TRI10 | Regulatory gene | No TRIs | |
| TRI9 | Unknown | Not determined. | |
| TRI11 | Isotrichodermin 15‐oxygenase | Isotrichodermin | |
| TRI12 | TRI efflux pump | No TRIs | |
| TRI13 | Calonectrin 4‐oxygenase | 4‐deoxy T‐2 toxin, 8‐hydroxycalonectrin, 8‐hydroxy‐3‐decalonectrin | |
| TRI14 | Virulence factor | T‐2 toxin | |
| TRI1 | C‐8 or C‐7,8 oxygenase | 4,15‐DAS | |
| TRI16 | C‐8 acyltransferase | Neosolaniol | |
| TRI101 | C‐3 acyltransferase | Isotrichodermol | |
| FUMs | FUM21 | Cys‐6 transcription factor | No FUMs |
| FUM1 | PKS | No FUMs | |
| FUM6 | Cytochrome P450 monooxygenase and reductase | No FUMs | |
| FUM7 | Alcohol dehydrogenase | Tetradehydro‐FB | |
| FUM8 | α‐Oxoamine synthase | No FUMs | |
| FUM3 | Dioxygenase | FB2, FB4 | |
| FUM10 | Acyl‐CoA synthetase/acyl‐protein synthetase | Hydrolyzed FB3, hydrolyzed FB4 | |
| FUM11 | Tricarboxyllic acid transporter | FB1, FB2, FB3, FB4 | |
| Half‐hydrolyzed FB1, FB2, FB3, FB4 | |||
| Keto half‐hydrolyzed FB1, FB2, FB3, FB4 | |||
| FUM2 | Cytochrome P450 monooxygenase | FB2, FB4 | |
| FUM20 | Unknown | Not determined | |
| FUM13 | Short‐chain dehydrogenase/reductase | 3‐keto FB3, 3‐keto FB4 | |
| FUM14 | Nonribosomal peptide synthase (peptidyl and condensation domains) | Hydrolyzed FB3, hydrolyzed FB4 | |
| FUM15 | Cytochrome P450 monooxygenase | No effect | |
| FUM16 | Acyl‐CoA synthetase/acyl‐protein synthetase | No effect | |
| FUM17 | Ceramide synthase | No effect | |
| FUM18 | Ceramide synthase | No effect | |
| FUM19 | ABC transporter | Increased ratio FB1:FB3 | |
| ZEA | PKS13 | PKS | No ZEA |
| PKS4 | PKS | No ZEA | |
| ZEB1 | Alcohol oxidase | β‐ZEL | |
| ZEB2 | Transcription factor | No ZEA |
The production profiles of Fusarium mycotoxins were determined by liquid chromatography–mass spectrometry analysis of extracts and filtrates of Fusarium mycotoxin biosynthesis gene mutants. FUMs, fumonisins; PKS, polyketide synthase; TAS, 3,4,15‐triacetoxyscirpenol; TRIs, trichothecenes; ZEA, zearalenone. Data were extracted from Alexander and colleagues 21 , 23 , 24 .
DON (also called vomitoxin) is one of the mycotoxins linked to FHB, and it is synthesized in F. graminearum, F. culmorum, and so on. The formal chemical name of DON is 3α,7α,15‐trihydroxy‐12,13‐epoxymonospora‐9‐ene‐8‐one 25 . It mainly contaminates cereal crops, such as maize, wheat, rice, and barley, as well as some cash crops. All over the world, DON contamination causes economic losses of billions of dollars each year. The Panel on Contaminants in the Food Chain, which is part of the European Food Safety Authority, examined 26,613 cereal samples from 21 European countries and found that the DON contamination rate was close to 50% 26 . Yan et al. collected 579 wheat samples and 606 maize samples from the major wheat‐ and maize‐producing provinces in China and determined the co‐occurrence of type‐B TRIs; all of the wheat samples showed positive results for DON, whereas 99.83% of the maize samples were DON‐positive 27 . In humans and animals, DON typically causes diarrhea, vomiting, and gastrointestinal inflammation. The chronic consumption caused by DON can result in immune‐suppressive diseases, growth impairment, and abnormalities of the reproduction and nervous systems 28 . The FAO and World Health Organization (WHO) first identified DON as a highly hazardous food contaminant in the 1970s 25 .
By binding to the 60S ribosomal subunit, DON can disrupt the action of peptidyl transferase and inhibit protein synthesis 18 , 28 . Moreover, DON can modulate mitogen‐activated protein kinase (MAPK) activity, which includes extracellular signal‐regulated kinase (ERK), c‐Jun N‐terminal kinase (JNK), and p38 mitogen‐activated protein kinase (p38), via the “ribotoxic stress response” by rapidly activating double‐stranded RNA‐associated protein kinase (PKR) and hematopoietic cell kinase (Hck 29 ; Figure 5A). DON‐induced MAPK activation can mediate the upregulation of transcription factors (nuclear factor kappa‐light‐chain‐enhancer of activated B cells [NF‐κB], activating protein 1 [AP‐1], and CCAAT/enhancer‐binding protein [C/EBP]) to promote the expression of proinflammatory cytokines, such as interleukin‐6 (IL‐6), IL‐1β, and tumor necrosis factor‐α (TNF‐α), which cause inflammation 28 , 30 . DON causes oxidative stress by inhibiting the expression of glutathione (GSH), superoxide dismutase (SOD), catalase (CAT), and GSH peroxidase (GSH‐PX), which engage in the repair of oxidative stress‐induced damage, decreasing total antioxidant capacity and glutathione S‐transferase levels 31 . However, DON can also induce nuclear factor‐erythroid 2‐related factor 2 (Nrf2) and heme oxygenase‐1 (HO‐1) activation to remove excessive reactive oxygen species (ROS). These results show that DON not only elicits oxidative stress but also impedes its occurrence. The potential impact of DON may vary depending on the dosage, although the precise mechanism remains incompletely elucidated 32 .
Figure 5.
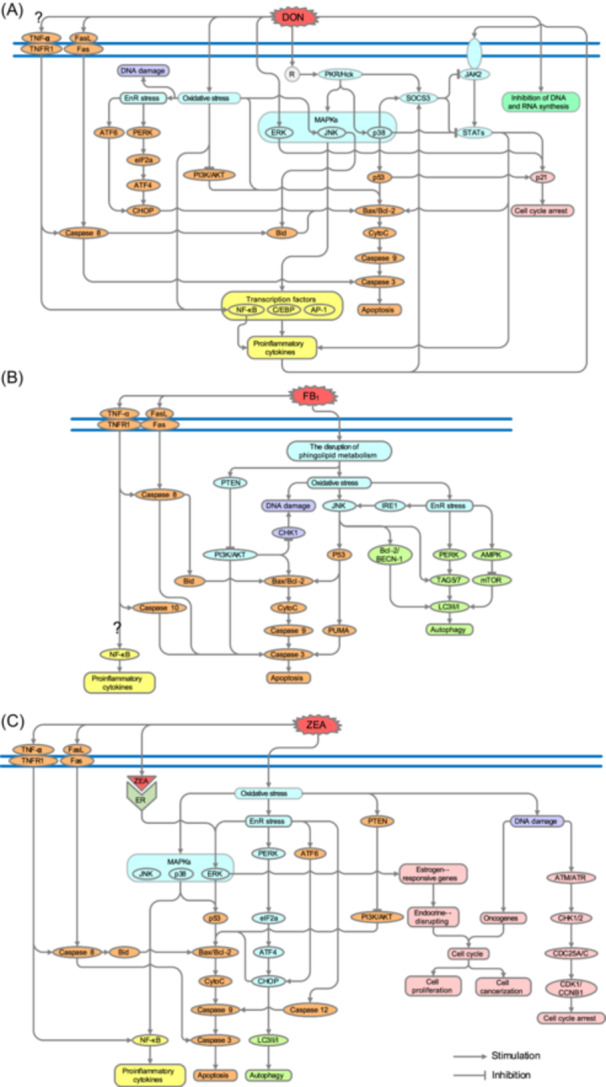
The toxicological mechanisms of the main Fusarium mycotoxins. (A) DON‐induced oxidative stress can cause DNA damage, EnR stress, and the suppression of the PI3K/AKT pathway. Consequently, it can lead to a decrease in Bcl‐2 gene expression, an increase in Bax gene expression, and elevated levels of CytoC expression. It enhances the upregulated expression of Caspase 9 and Caspase 3 and induces apoptosis. DON can efficiently induce the activation of MAPKs through a mechanism referred to as the “ribotoxic stress response” and hijack the JAK2/STAT‐3 pathway to induce apoptosis, proinflammatory cytokine upregulation, and cell cycle arrest. DON also activates Caspase 8 to increase expression of Caspase 3 by the tumor necrosis factor‐α and Fas/FasL signaling pathways, leading to apoptosis. (B) FB1 disrupts sphingolipid metabolism, causing DNA damage and apoptosis through the PTEN/PI3K/AKT signaling pathway as well as oxidative stress, which further entrenches DNA damage and apoptosis and induces EnR stress. FB1‐induced EnR stress not only causes apoptosis by the JNK/p53/PUMA/Caspase 3 pathway but also leads to autophagy via signaling pathways mediated by IRE1, PERK, and AMPK. IRE1 can activate JNK, which results in the subsequent elevation of BECN1, ATG5, and ATG7 and the conversion of LC3‐I. PERK induces the expression of ATG5 and ATG7 to form the ATG5–ATG7 complex and promotes the conversion of LC3‐I into LC3‐II. Under EnR stress, AMPK inhibits the anabolic process of mTOR, thereby increasing the expression of autophagy‐related genes and triggering autophagy. (C) ZEA and its derivatives can bind to ERs to elicit estrogen‐like effects and promote cellular proliferation. ZEA‐induced DNA damage can upregulate ATM and ATR expression and activate DNA damage checkpoints CHK1 and CHK2 to repair DNA damage. Next, the expression levels of CDC25A and CDC25C are upregulated, which promotes the expression of CCNB1 and CDK1, thus preventing the cell cycle from exiting the G2/M phase. Moreover, ZEA can induce apoptosis and autophagy similar to FB1 and DON. Question marks indicate indeterminate roles. EnR, endoplasmic reticulum; MAPK, mitogen‐activated protein kinase.
The relationship between oxidative stress and inflammation is interrelated and interdependent. Specifically, the products of oxidative stress (primarily ROS) have been shown to increase proinflammatory responses. Additionally, inflammatory cells release significant amounts of ROS at sites of inflammation, exacerbating oxidative damage 33 . In most cell types, mitochondria are not only the largest contributors to intracellular ROS production but also the targets of cellular ROS. A feed‐forward detrimental cycle exists between the generation of ROS and mitochondrial damage, wherein ROS‐damaged mitochondria undergo dysfunctionality, subsequently leading to an exacerbation in intracellular ROS production 34 . In the case of oxidative stress, damaged mitochondria show a decrease in mitochondrial membrane potential, which is closely related to apoptosis 28 , 32 . The B‐cell lymphoma 2 (Bcl‐2) gene family regulates apoptosis through the mitochondrial pathway. Among this gene family, the Bcl‐2‐associated X (Bax) gene and Bcl‐2 antagonist/killer‐1 (Bak‐1) gene are important proapoptotic genes, while Bcl‐2 is an antiapoptotic gene. The augmented Bax/Bcl‐2 ratio serves as a pivotal indicator of the initiation of apoptosis. Caspases (cysteine‐aspartic proteases, cysteine aspartases, or cysteine‐dependent aspartate‐directed proteases), a group of protease enzymes, are crucial players in the process of apoptosis, with Caspase 3 being known as the executor of apoptosis 35 . DON‐induced oxidative stress results in oxidation and damage of DNA and lipids through the elevation of ROS levels, finally leading to apoptosis through the mitochondrial pathway via activation of the JNK pathway, which increases the Bax/Bcl‐2 ratio and induces the conversion of Caspase 9 into Caspase 3. In addition, this process triggers proinflammatory cytokine expression by upregulating NF‐κB 32 , 36 , 37 . Moreover, DON can also induce apoptosis by regulating the Janus kinase 2/signal transducers and activators of transcription (JAK2/STAT) pathway and inhibiting the phosphatidylinositol 3‐kinase/threonine kinase (PI3K/AKT) pathway, the latter of which is important in inhibiting apoptosis because it regulates downstream effector molecules (e.g., suppressing Bax translocation) 36 , 38 , 39 , 40 . In addition, DON can disturb normal cell cycle progression by upregulating ERK and JAK2/STAT to promote cyclin‐dependent kinase inhibitor p21 expression 40 . Perturbations in endoplasmic reticulum (EnR) function resulting from enhanced protein synthesis or the accumulation of misfolded proteins give rise to a condition known as EnR stress 41 . The generation of ROS can elicit an elevation in the EnR stress level, while EnR stress can induce the production of ROS in the EnR and mitochondria 42 . EnR stress can induce cellular death, including apoptosis and autophagy, through three classical signaling pathways (protein kinase RNA‐like endoplasmic reticulum kinase [PERK], activating transcription factor 6 [ATF6], and the inositol‐requiring protein 1 [IRE1]‐mediated signaling pathway). In EnR stress‐induced apoptosis, the C/EBP homologous protein (CHOP) transcription factor plays a pivotal role in the PERK, ATF6, and IRE1 signaling pathways 43 . To date, studies have demonstrated only that 3ADON has the capability to induce apoptosis in a mouse liver by inducing EnR stress via the IRE1 pathway 44 . No similar study of DON yet exists. The DON‐activated death receptor pathway of apoptosis includes the TNF‐induced model and the Fas/FasL‐mediated model (Fas, the first apoptosis signal, is also known as Apo‐1 or CD95; FasL, Fas ligand), both of which are linked to TNF receptor (TNFR) family receptors that are connected to extrinsic signals. Although the exact mechanism is not fully understood, DON has demonstrated an ability to hinder DNA and RNA synthesis 18 , 28 . Moreover, You et al. 32 raised the presumption that DON and T‐2 could achieve the “immune evasion” process to actively evade immune surveillance by immune cells. A recent study showed that T‐2 could initiate the “immune evasion” process by activating the signaling pathway involving the programed cell death protein‐1/programmed cell death‐ligand 1 45 . For DON, however, there is a dearth of related studies.
FUMs
FUMs are a class of long‐chain amino polyalcohols that are primarily synthesized by species in the Fusarium fujikuroi species complex, especially in Fusarium verticillioides and F. fujikuroi (Figure 2). FUMs occur primarily in cereals (rice, wheat, barley, maize, rye, oat, and millet). Over 28 FUM homologs are known, and particular emphasis has been given to the B series, notably FUM B1 (FB1), FB2, and FB3 11 . FB1 is composed of a diester comprising propane‐1,2,3‐tricarboxylic acids (TCA) and 2‐amino‐12,16‐dime thyl‐3,5,10,14,15‐pentahydroxyleicosane, where hydroxyl groups at the C‐14 and C‐15 positions interact with the carboxyl groups of TCA to form an ester. Moreover, FB2 and FB3 can be regarded as the C‐5 and C‐10 dehydroxy analogs of FB1 19 (Figure 3B). FUMs induce a range of deleterious effects on organisms, encompassing carcinogenicity, cytotoxicity, hepatotoxicity, immunotoxicity, nephrotoxicity, neurotoxicity, and reproductive toxicity 46 .
In F. verticillioides, the biosynthesis of FUMs necessitates a sophisticated gene cluster spanning 42 kb with 17 coregulated genes (Figure 4A). Except for FUM20, the FUM production profiles of FUM gene deletion or disruption mutants were ascertained using liquid chromatography–mass spectrometry (LC‐MS) analysis (Table 1). FUM1, which encodes a polyketide synthase (PKS), is the key gene in FUM biosynthesis. In the first step, the FUM1 protein (Fum1p) facilitates the condensation of 1 acetyl coenzyme A (acetyl‐CoA), 8 malonyl‐CoAs, and 2 S‐adenosyl methionines (SAMs) to generate a linear 18‐carbon‐long polyketide. In the second step, Fum8p catalyzes the condensation between the linear polyketide and alanine, resulting in a 3‐keto intermediate with a 20‐carbon chain. In the third step, Fum6p is responsible for catalyzing the hydroxylation process of the polyketide‐amino acid condensation product at C‐14 and C‐15. Then, the carbonyl group at C‐3 is reduced into an alcohol group by Fum13p. The addition of a hydroxyl group to the C‐10 carbon is catalyzed by Fum2p. The catalysis of esterification, leading to the hydroxylation at the C‐14 and C‐15 positions of the FUM backbone, is facilitated by Fum14p. In addition, the involvement of Fum7p, Fum10p, and Fum11p is also evident in the biosynthesis of the tricarballylate portion. The final step is the hydroxylation of the FUM backbone at C‐5 by Fum3p (Figure 4C). Fum21p, a transcription regulator with a Zn(II)2Cys6 DNA‐binding domain, can positively regulate the gene expression of FUM1 and FUM8 and is also required for FUM synthesis 21 .
FB1, the most typical FUM, has a structure similar to that of sphingolipids (SLs), and it has the capability to act as an inhibitor of ceramide synthase (CerS). Inhibition of CerS interferes with SL metabolism, causing the accumulation of free sphinganine (Sa) and sphinganine‐1‐phosphates (Sa‐1‐P) in cells, alterations in complex SLs, and reduced levels of ceramides 46 . FB1 and its generated toxic products (Sa, Sa‐1‐P) induce oxidative stress by exacerbating peroxide production (ROS, H2O2, lipid peroxide, and lipid oxidation end products) and inhibiting the activity of antioxidants (SOD, CAT, GSH‐PX, and GSH 46 ; Figure 5B). FB1‐induced oxidative stress can induce JNK phosphorylation, activate P53 signaling, and upregulate the expression of proapoptotic factors (P53‐upregulated modulator of apoptosis [PUMA] and Caspase 3) to cause apoptosis 41 , 47 . FB1‐induced EnR stress leads to apoptosis through the JNK/p53/PUMA/Caspase 3 pathway and autophagy through the IRE1/JNK pathway, which releases Beclin‐1 (BECN1) from the Bcl‐2‐BECN‐1 interaction to promote the conversion of microtubule‐associated protein 1 light chain 3 (LC3)‐I into LC3‐II 47 . In addition, FB1‐induced EnR stress can induce autophagy by the PERK pathway and the AMP‐dependent protein kinase (AMPK) pathway 47 , 48 . FB1 exposure can also induce apoptosis by regulating the phosphatase and tensin homolog (PTEN)/PI3K/AKT signaling pathway via disruption in lipid raft formation 49 . Moreover, FB1 exerts an epigenetic influence on the PTEN/PI3K/AKT signaling pathway to enhance DNA damage by inhibiting checkpoint kinase 1 (CHK1) activity through phosphorylation of its Ser280 residue, thereby impeding the repair process for damaged DNA 50 . It has also been demonstrated that the apoptotic pathway of FB1 is linked to death receptor pathways, including the TNF pathway and the Fas pathway 46 . In the TNF pathway, the function of NF‐κB is complex. Gopee et al. 51 suggested that FB1‐induced apoptosis involved the activation of Caspase 3 in pig kidney epithelial cells (LLC‐PK1), which was correlated with the suppression of NF‐κB. However, Chen et al. 52 suggested that FB1 treatment resulted in the upregulation of both Caspase 3 and NF‐κB in pig kidney (PK‐15) cells.
ZEA
ZEA, formerly known as F‐2 toxin, is a resorcylic acid lactone (Figure 3C) that is synthesized by some members of the F. sambucinum species complex, the F. incarnatum‐equiseti species complex, and the F. fujikuroi species complex (Figure 2), such as F. graminearum, F. culmorum, F. equiseti, and F. verticillioides 11 . The toxicity of ZEA encompasses various dimensions, including alimentary canal toxicity, endocrine interference, carcinogenicity, genotoxicity, hepatotoxicity, immunotoxicity, and reproductive toxicity. ZEA predominantly contaminates grains, including maize, wheat, rice, barley, sorghum, soybean, oat, and their products 53 .
In F. graminearum, the ZEA biosynthesis gene cluster contains four genes: PKS13, PKS4, ZEB1, and ZEB2 (Figure 4A). Disruption of PKS13, PKS4, or ZEB2 can result in a permanent halt in ZEA production, and the ZEB1 deletion mutant produces the ZEA derivative β‐zearalenol (β‐ZEL; Table 1). PKS4 can catalyze the synthesis of the hexaketide chain using one acetyl‐CoA and five malonyl‐CoA units. Then, the hexaketide is transferred to the nonreducing PKS13 to form nonaketide after completing three condensations. The nonaketide subsequently undergoes two successive intramolecular cyclization reactions, leading to the formation of an aromatic ring and a macrolide ring structure containing a lactone bond. Finally, β‐ZEL is transformed to ZEA by ZEB1, which facilitates the transformation of the hydroxyl group on the macrolide into the ketone group 23 (Figure 4D).
ZEA and its metabolites have a three‐dimensional (3D) structural similarity to estradiol and can exert estrogen‐like effects. Estrogen regulates physiological processes via estrogen receptors (ERs), which are capable of initiating many signaling pathways. At low concentrations, ZEA usually induces the proliferation of cells through estrogen‐like effects and carcinogenic properties 54 . ZEA and its metabolites can occupy and activate ERs and then mediate the expression of estrogen‐responsive genes through the ERK signaling pathway 55 (Figure 5C). The modulation of physiological estrogen responses, such as endocrine disruption and cell proliferation, is attributed to the expression of genes regulated by estrogen 56 . The DNA damage caused by ZEA might lead to mutations or chromosome abnormalities, which can disturb the progression of the cell cycle and cause cell proliferation or cell cancerization 57 , 58 . Furthermore, ZEA downregulates the expression of tumor suppressor genes and upregulates the expression of oncogenes in TM3 cells, potentially promoting the conversion of normal cells into malignant cells 59 . Similarly, a change in oncogene expression might cause cell proliferation. Moreover, DNA damage caused by ZEA can also lead to cell cycle arrest. DNA damage causes the upregulated expression of ataxia‐telangiectasia mutated serine/threonine kinase (ATM) and ataxia telangiectasia and Rad3‐related protein (ATR), which activate CHK1 and CHK2, and cells begin to repair the damage. However, ZEA‐exposed cells undergo arrest in the G2/M phase, during which there are upregulated expression of cell division cycle 25 phosphatases (CDC25) A and CDC25C. These two proteins subsequently enhance the expression of cyclin B1 (CCNB1) and cyclin‐dependent kinase 1 (CDK1), thereby preventing exit from the G2/M phase of the cell cycle. The arrest of cell cycle progression triggers a halt in DNA replication and consequently inhibits cellular proliferation 60 , 61 , 62 . At high concentrations, ZEA causes mitochondrial dysfunction, EnR stress, apoptosis, and autophagy. ZEA reduces the protein expression of Nrf2 and HO‐1 to further induce oxidative stress and cause cell apoptosis via the p38, JNK, and ERK MAPK pathways 63 , 64 , 65 . Bai et al. found that ZEA can induce apoptosis by modulating EnR stress though the PERK and ATF6 signaling pathways in porcine trophectoderm cells 66 . In addition to the classical signaling pathways, ZEA induces apoptosis through the ERK/p53/Caspase 3 signaling pathway and the Caspase 12 signaling pathway activated by Ca2+ release from the EnR 67 , 68 . ZEA‐induced EnR stress also causes autophagy through the PERK pathway 69 . Moreover, ZEA can cause apoptosis by activating death receptor pathways and inhibiting the PI3K/AKT pathway and can induce the expression of proinflammatory cytokines through the TNF/NF‐κB pathway and MAPK/NF‐κB pathway 70 , 71 , 72 , 73 .
ZEA can be biotransformed in the liver and bacterial gut flora of mammals by hydroxysteroid dehydrogenases (HSD). There are 4 reductive metabolites of ZEA in the reductive phase‐I metabolism process: α‐ZEL, β‐ZEL, α‐zearalanol (α‐ZAL, also known as zeranol), and β‐ZAL (Figure 3C). 3α‐ and 3β‐HSD can catalyze the hydroxylation of ZEA, resulting in stereoisomeric compounds α‐ and β‐ZEL, respectively. Furthermore, with the saturation of a double bond, α‐ and β‐ZEL can be further reduced into α‐ and β‐ZAL, respectively (Figure 4E). α‐ZEL and α‐ZAL show higher xenoestrogenic effects than ZEA, while β‐ZEL and β‐ZAL are just the opposite. α‐ZAL has been extensively used as a growth enhancer to augment the fattening rates of cattle. Since 1985, the application of α‐ZAL has been banned in the European Union. 74
CC AND FUSARIUM MYCOTOXIN CONTAMINATION
On the basis of the Sixth Assessment Report of the Intergovernmental Panel on Climate Change, it is projected that global temperatures will continue to rise, while atmospheric concentrations of carbon dioxide ([CO2]) are anticipated to undergo a twofold or even threefold increase within the next 25–50 years 75 . Adverse CC can lead to more frequent occurrences of extreme weather events, such as heat waves, cold waves, heavy rainfall, and drought. CC is a global challenge and is forecasted to have significant impacts on food security by influencing crop growth, the occurrence of pests and diseases, and mycotoxin contamination. CC may lead to transformations in the spatial distribution and manifestation of mycotoxigenic fungi, thereby causing alterations in both the geographical distribution and the occurrence pattern of mycotoxins. Studies have shown that temperature, water activity (a w), [CO2], or a combination of the three variables can also impact the growth, proliferation, and production of mycotoxins from mycotoxigenic fungi 76 (Figure 6). Although pathogenic Fusarium species are important producers of mycotoxins, there is no clear correlation, in general, between disease severity and mycotoxin contamination 77 . At Bottelare village, East Flanders province, Belgium, a positive correlation between FHB severity and DON content was observed during the growing seasons of 2001–2002 and 2002–2003; however, this relationship was not evident in the 2003–2004 season 78 .
Figure 6.
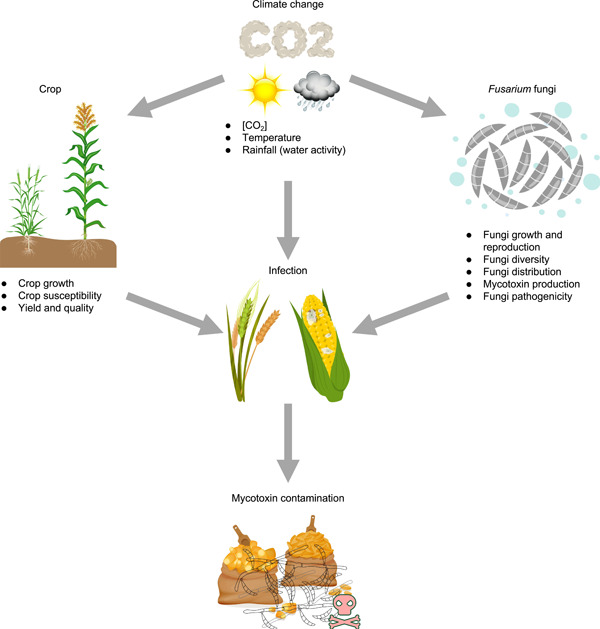
The impact of climate change on Fusarium mycotoxin contamination. Temperature, rainfall (or water activity), and carbon dioxide concentration ([CO2]) are the main known climate drivers affecting Fusarium mycotoxin contamination. Climate factors not only influence the growth and development of crops and Fusarium fungi but also affect the infection process of Fusarium fungi through changes in crop susceptibility, virulence of Fusarium fungi, and environmental factors at critical stages of infection.
Temperature and rainfall
Although FUMs, DON, and ZEA have been detected all over the world, because of the different optimal production temperatures and a w values of Fusarium fungi (Table 2), the contaminants have clear differences in geographical distribution. FUMs are the main Fusarium mycotoxin contaminants in southern Europe, the Americas, the Middle East, Africa, and south and southeast Asia. DON contamination occurs mostly in northern, central, and eastern Europe, northern and central America, South Africa, and East Asia. Compared to those of FUMs and DON, the occurrence rate of ZEA is lower and is only over 50% in East Asia and sub‐Saharan Africa 79 .
Table 2.
The optimal temperature and a w of Fusarium mycotoxin production.
| Fusarium verticillioides | Fusarium proliferatum | Fusarium graminearum | Fusarium culmorum | |||||
|---|---|---|---|---|---|---|---|---|
| Mycotoxin | Temperature (°C) | a w | Temperature (°C) | a w | Temperature (°C) | a w | Temperature (°C) | a w |
| FUMs | 15–32 | >0.940 | 13–25 | >0.960 | ||||
| DON | 15–35 | >0.980 | 20–30 | >0.975 | ||||
| ZEA | 25–30 | >0.980 | ||||||
The optimal temperature and a w of FUMs and ZEA production were detected on Fusarium‐infected maize grain. The optimal temperature and a w of DON production were detected on Fusarium‐infected wheat grain. Data were extracted from Sanchis and colleagues 80 , 81 . a w, water activity; DON, deoxynivalenol.
Against the backdrop of global warming, regions currently characterized by cooler climates will witness the prevalence of toxigenic fungi that show optimal growth and mycotoxin production under higher‐temperature conditions. Conversely, areas already experiencing hot temperatures might observe a decrease in the occurrence of such fungi. With rising temperatures, lower crop yield and quality will occur in some regions that are currently considered warm. This might result in a reduction in total mycotoxin production due to the reduced crop quantity. However, due to the lower quality of crops, the mycotoxin content per unit weight of crops might increase 82 . Temperature is closely related to latitude and altitude. In most regions where maize is grown, FUM contamination tends to be higher in areas with lower latitudes and altitudes due to relatively warmer conditions compared to regions with higher latitudes or altitudes 83 . In the United States, the FUM risk is higher in Texas and the southeastern states than in the central states 84 . Shelby et al. 85 discovered a significant inverse relationship between latitude and FUM concentration in the United States. A similar pattern can also be found in Asia, north of the Tropic of Cancer. In the low‐altitude maize production areas of central and south America, as well as southeast Asia, FUMs emerge as a significant risk factor. In Guatemala, a survey of maize samples gathered from fields between 2000 and 2003 showed that lowland maize exhibited significantly higher levels of FB1 than highland maize did 86 . In Europe, the risk of FUM contamination is higher in Italy, Spain, and southern France. In Africa, all maize‐producing areas are at risk for FUMs, with severity depending on altitude 84 . A survey in Uganda found that the FUM contamination of maize was widespread and that maize from high altitudes showed the most significantly elevated levels of FUM content 87 . DON contamination is also sensitive to temperature changes. In northwestern European countries, according to the prediction model, the flowering and full maturation of wheat will advance with the relative increase in temperature, and most regions will show a substantial rise in DON contamination in the 2040s 88 .
Although numerous studies have demonstrated that Fusarium mycotoxin production requires high a w in vitro (Table 2) 80 , 81 , none have demonstrated monotonic relationships between rainfall and Fusarium mycotoxin contamination across different environments. In Ontario, Canada, a cool maize‐growing region, FUMs are only present in drought‐stressed fields 89 . In the United States, there is a significant negative correlation between June rainfall and FUM content at multiple locations 85 . Akello et al. 90 studied the FUM content of cereals in Zimbabwe during 2015 and 2017 and found that FUM contamination was higher in wet years than in dry years. According to existing meteorological data in the Philippines, Salvacion et al. 91 built a risk model of FUM contamination on corn using a fuzzy logic methodology. Due to the increased rainfall, they argued that a substantial proportion of the Philippines might be at a very high risk under prevailing circumstances, as well as under the anticipated CC scenarios for 2050. During the period 2012–2021, in both Serbia and Croatia, the highest mean contents of DON and ZEA in maize were observed in 2014, which might be related to the extreme precipitation during that year 92 . A 10‐year (2008–2017) global survey showed that maize harvested in central and southern Europe exhibited elevated levels of DON and ZEA concentrations in 2014, which corresponded to higher rainfall in July 2014. In the primary maize‐growing regions of China, DON and ZEA levels were relatively low in 2013, which might be related to the decrease in precipitation during August and September of that year 79 .
The period around flowering is considered to be the pivotal stage for Fusarium infection of cereals. Therefore, many studies have been dedicated to predicting the concentrations of Fusarium mycotoxins by the weather variables of the period around flowering, especially temperature and rainfall (Table 3). Campa et al. 93 built a model to predict the FUM concentration of maize by using data gathered from Argentina and the Philippines. The model showed that weather was the major variable in total FUM concentration; the temperature and precipitation of the four periods around silking were determined to be the key factors in the FUM concentration. Hooker et al. 94 identified a comprehensive set of weather variables and their temporal patterns for the prediction of DON contamination in mature wheat grain in Canada. They found that contamination showed a significant correlation with weather conditions during three pivotal periods around the heading stage. In the first period, 4–7 days before heading, DON generally decreased with the number of days with temperatures below 10°C and increased with the number of days experiencing rainfall exceeding 5 mm. In the second period, 3–6 days after heading, DON levels showed an upward trend in correlation with an increase in the number of days with rainfall exceeding 3 mm and a downward trend when exposed to temperatures exceeding 32°C. In the third period, 7–10 days after heading, DON increased with an increase in days with rainfall exceeding 3 mm. In Schleswig‐Holstein, northern Germany, Birr et al. 95 found that there were significant relationships between the two weather variables (cumulative precipitation and average temperature during the period of wheat flowering) and the concentrations of ZEA in wheat grain at harvest. Based on this finding, they derived weather‐based forecasting models for predicting ZEA levels in wheat grain during the harvest stage for various Fusarium‐susceptible wheat cultivars. Joo et al. 96 constructed a model to assess the influence of CC on ZEA contamination in rice grains cultivated in South Korea. The results indicated that increased temperature and relative humidity during the rice heading period and fluctuations in daily temperature throughout the harvest season can increase ZEA contamination in rice. The forecasts showed that ZEA contamination of rice could increase nationwide in both the 2030s and the 2050s, particularly in the western region of South Korea.
Table 3.
The conditions for predicting the impact of CC on Fusarium mycotoxin contamination.
| Mycotoxin | Crop | Critical period | Increased danger | Decreased danger | Ref. |
|---|---|---|---|---|---|
| FUMs | Maize |
|
RAIN >2 mma |
TMIN < 15 °Cb, TMAX > 34 °C |
[91] |
|
TMAX > 34°C | ||||
|
TMAX > 34°C |
RAIN >2 mm, TMIN < 15 °C |
|||
|
RAIN >2 mm | ||||
| DON | Wheat |
|
RAIN >5 mm | TMIN < 10 °C | [92] |
|
RAIN >3 mm | TMAX > 32 °C | |||
|
RAIN >3 mm | ||||
| ZEA | Wheat | Flowering period | The higher cumulative precipitation and average temperature | [93] | |
| Rice |
|
The higher average temperature and humidity | [94] | ||
|
The higher daily temperature changes |
The risk of Fusarium mycotoxin contamination will increase with the number of days experiencing rainfall exceeding 2 mm. bThe risk of Fusarium mycotoxin contamination will decrease with the number of days when the minimum temperature is below 15 °C.
[CO2] level
Although the impacts of elevated [CO2] on crops have been studied in depth 97 , there remains a scarcity of studies examining the responses of plant disease to increased [CO2]. Furthermore, research that specifically focuses on mycotoxins in plants is even more limited. Currently, the effects of [CO2] on Fusarium mycotoxin contamination have mainly been studied in laboratories; large‐scale field surveys have not yet been conducted. Elevated [CO2] can increase plant susceptibility to Fusarium species attacks. Rising [CO2] was found to increase F. verticillioides proliferation in maize with no change in FUM levels. This result indicates a decrease in the production of FUMs per unit of pathogen. Following F. verticillioides infection at elevated [CO2], the suppression of maize 13‐lipoxygenase and jasmonic acid production was correlated with a decrease in terpenoid phytoalexins and an increase in susceptibility to the pathogen, while a reduction in 9‐lipoxygenase, previously proposed to enhance mycotoxin production, was responsible for reduced FUMs per unit fungal biomass 98 . Further research showed that concurrent elevated [CO2] and drought stress significantly augmented the susceptibility of maize to F. verticillioides infection, consequently leading to an escalated contamination of FUMs. However, the negative impacts of drought on the accumulation of maize phytohormones and metabolites were not mitigated by elevated [CO2], and there was still no observed increase in FUMs per unit fungal biomass. Therefore, it is likely that the escalation in FUM contamination can be attributed to the greater F. verticillioides biomass 99 . F. graminearum infection produced similar results. Hay et al. 100 found that elevated [CO2] can significantly increase F. graminearum biomass and DON accumulation in maize, but the DON per unit fungal biomass was unaffected. For wheat, it was a different story. In the F. culmorum single‐floret inoculation treatment, the concentration of DON was significantly increased under elevated [CO2]. This result suggests that the DON content is not directly related to the level of infection with F. culmorum 101 . In addition, acclimatization to elevated [CO2] can impact the mycotoxin production of Fusarium fungi. A recent in vitro study demonstrated that, under elevated [CO2] conditions, F. sporotrichioides had a greater ability to produce T‐2 and HT‐2 after 10 subculture generations than in the initial subculture of the strain 102 .
DETECTION OF FUSARIUM MYCOTOXINS
Typically, analysis methods for mycotoxins require three major steps: extraction, cleanup, and detection 103 . Mycotoxins can be detected using various techniques, mainly chromatographic methods, immunological methods, and biosensor technologies (Table 4). However, each approach has advantages and disadvantages. The selection of a particular method is contingent upon the specific detection requirements 20 .
Table 4.
The detection methods of main Fusarium mycotoxins.
| Detection method | Time (min) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mycotoxin | Chromatography | Immunoassay | Biosensor | Matrix | Sample preparation method | Preparation | Assay | LOD (ppb) | Specificity | Ref. |
| FB1 | LC‐MS/MS | Corn | SLE | 92 | 21 | 8 | [104] | |||
| Corn | SLE | 120 | 30 | 100 | [105] | |||||
| Corn | SLE | 100 | 15 | 3 | [106] | |||||
| Peanut | SLE | 120 | 30 | 5 | [105] | |||||
| Pistachio | SLE | 120 | 30 | 10 | [105] | |||||
| Wheat | SLE | 120 | 30 | 10 | [105] | |||||
| Wheat | SLE | 100 | 15 | 0.5 | [106] | |||||
| Raisin | SLE | 120 | 30 | 5 | [105] | |||||
| HPLC‐MS/MS | Wheat flour | SLE | >65 | 30 | 12 | [107] | ||||
| Wheat flour | SPE | >30 | 0.01 | [108] | ||||||
| Corn | SPE | >11 | 25 | 0.64 | [109] | |||||
| HPLC‐FLD | Corn | SLE | >15 | 50 | [110] | |||||
| Tortilla, masa, corn | SLE | >45 | 25 | [111] | ||||||
|
Canned sweet corn, fresh sweet corn, corn grits, corn flour, cornflakes |
SLE | 180 | 25 | 29.2 | [112] | |||||
| Cereal foods | IAC | 20 | 14.6 | [113] | ||||||
| UPLC‐MS/MS | Alpinia oxyphylla | SLE | 35 | 32 | 0.2 | [114] | ||||
| Corn | QuEChERS | 60 | 25 | 6.3 | [115] | |||||
| ELISA | Corn | SLE | 15 | 1110 | 8 | 224% and 73% CRs with FB2 and FB3 | [110] | |||
|
Corn and corn related samples |
SLE | 50 | 960 | 1 | 5% CR with T‐2 | [116] | ||||
| Corn | SPE | 120 | 0.19 | 6.89% and 2.93% CRs with FB2 and FB3 | [117] | |||||
| Corn | SLE | 40 | 915 | 1.15 | 60.4% CR with FB2 | [118] | ||||
| Corn, feedstuff, wheat | SLE | 10 | 730 | 1.18 | The negligible CRs with FB2, OTA, ZEA, DON, and AFB1 | [119] | ||||
| LFI | Corn | SLE | 15 | 10 | 25 | No CRs with ZEA, DON, OTA, AFB1, and FB1 | [120] | |||
| Corn | SLE | 30 | 10 | 0.5 | No CRs with AFM1, DON, FB2, T‐2, and FB3 | [121] | ||||
| Feed | SLE | 30 | 10 | 1.94 | AFM1, DON, FB2, T‐2, and FB3 did not interfere with the detection of FB1 | [121] | ||||
| Corn, wheat | SLE | 15 | 5 | 20 | AFB1, ZEA, and OTA did not interfere with the detection of FB1 | [122] | ||||
| Chinese traditional medicine | SLE | 25 | 5 | 5 | No CRs with AFB1, ZEA, and OTA | [123] | ||||
| MIP‐ELISA | Corn | SLE | 1445 | 1.9 × 10−3 | The negligible CRs with FB2, AFB1, CIT, ZEA, and DON | [124] | ||||
| MIP‐EC biosensor | Corn | SLE | 6 | 8.89 × 10−6 | The negligible CRs with AFB1, CIT, DON, and ZEA | [125] | ||||
| MIP‐ECL biosensor | Milk, corn | LLE, SLE | 30 | 15 | 3.5 × 10−4 |
The negligible ECL signals of OTA, OTB, DON, CS, LAC, DA, and NE |
[126] | |||
| MIP‐PEC biosensor | Milk, corn | LLE, SLE | 30 | 20 | 4.7 × 10−3 | The negligible photocurrents of OTA, OTB, DON, ZEA, PAT, Glu, and starch | [127] | |||
| EC immunosensor | Corn | SLE | 20 | 40 | 4.2 | No response for DON | [128] | |||
| Corn | IAC | 180 | 0.002 | The peak currents caused by ZEA, OTA, and DON showed a comparable pattern to that observed in the control sample | [129] | |||||
| EC aptasensor | Beer | 10 | 2.6 × 10−4 | The peak current of OTA, ZEA, and AFB1 was higher significantly than FB1 | [130] | |||||
| Rice | SLE | 45 | 8.7 × 10−5 | The obvious differences of ECL signals between FB1 and AFB1, AFB2, DON, OTA, ZEA | [131] | |||||
| Colorimetric signal aptasensor | Corn, wheat | SLE | 25 | 30 | 0.024 | Effectively avoiding interferences of FB2, AFB1, DON, ZEA, and T‐2 | [132] | |||
| ECL aptasensor | Wheat | SLE | 0.27 | The obvious differences of ECL signal between FB1 and OTA, AFT, l‐cys, l‐Hcys | [133] | |||||
| DON | GC‐MS | Wheat | SPE | 90 | 24.2 | 3 | [134] | |||
| LC‐MS/MS | Corn | QuEChERS | 13 | 44 | 739 | [135] | ||||
| Corn | SLE | 120 | 30 | 50 | [105] | |||||
| Corn | SLE | 100 | 15 | 8 | [106] | |||||
| Peanut | SLE | 120 | 30 | 75 | [105] | |||||
| Pistachio | SLE | 120 | 30 | 50 | [105] | |||||
| Wheat | SLE | 120 | 30 | 20 | [105] | |||||
| Wheat | SLE | 100 | 15 | 35 | [106] | |||||
| Raisin | SLE | 120 | 30 | 9 | [105] | |||||
| UPLC‐MS/MS | Alpinia oxyphylla | SLE | 35 | 32 | 6 | [114] | ||||
| Corn | SPE | 26 | 9 | 0.1 | [136] | |||||
| Oat | SPE | 26 | 9 | 0.12 | [136] | |||||
| Corn | QuEChERS | 60 | 25 | 3.2 | [115] | |||||
| HPLC‐MS/MS | Wheat flour | SLE | >65 | 30 | 5.1 | [107] | ||||
| Corn | SPE | >11 | 25 | 0.29 | [109] | |||||
| HPLC‐FLD | Wheat | IAC | 21.7 | [137] | ||||||
| Corn | IAC | 14.08 | [137] | |||||||
| HPLC‐PDA | Cereal foods | IAC | 30 | 15.5 | [113] | |||||
| ELISA | Wheat | SLE | >15 | 45 | 0.62 | 4.7% CR with 3ADON | [138] | |||
| Cereals and cereal products | SLE | 20 | 790 | 4.9 | 5.7% CR with 3ADON | [139] | ||||
| Rice | SLE | 20 | 300 | 0.94 | [140] | |||||
| Rice, corn, flour, feed | SLE | 45 | 835 | 0.2 | 80.34%, 2.17%, and 2.74% CRs with 3ADON, 15ADON, and FUS‐X | [141] | ||||
| LFI | Corn, wheat | SLE | 7 | 5 | 100 |
No CRs for multianalysis of DON and ZEA |
[142] | |||
| Corn, wheat | SLE | 15 | 5 | 5 | AFB1, ZEA, and OTA did not interfere with the detection of DON | [122] | ||||
| Corn, wheat | SLE | >8 | 10 | 50 | 400%, 1.6%, and 4.3% CRs with 15ADON, 3ADON, and NIV | [143] | ||||
| Chinese traditional medicine | SLE | 25 | 5 | 5 | NO CRs with AFB1, ZEA, and OTA | [123] | ||||
| Rice, corn | SLE | 45 | 15 | 12.5 | 80.34%, 2.17%, and 2.74% CRs with 3ADON,15ADON, and FUS‐X | [141] | ||||
| SPR immunosensor | Corn, wheat | SLE | 45 | 20 | 3.26 | 16.2% CR with 15ADON | [144] | |||
| EC immunosensor | Wheat | SLE | 30 | 13 | 342.4 | 221% CR with 3ADON | [145] | |||
| MIP‐EC biosensor | Corn | SLE | 25 | 15 | 0.3 | Compared to OTA, FB1, FB2, NIV, and ZEA, MIP sensor showed higher recognition selectivity toward DON | [146] | |||
| Wheat flour | SLE | 40 | 6.5 | 0.021 | The ΔI after incubation in DON is exhibited higher than that in ascorbic acid, Cu2+, Glu, glutamic acid, OTA, K+, Na+, Mg2+, sucrose, and ZEA | [147] | ||||
| MIP‐SPR biosensor | Standard substance | 1 | 19% and 44% selectivity efficiencies with 3ADON and 15ADON | [148] | ||||||
| SERS aptasensor | Corn flour, peanut oil, pure milk | LLE, SLE | >40 | 40 | 3.2 × 10−5 | The obvious differences of SERS signal between DON and AFB1, OTA, FB1, T‐2, and ZEA | [149] | |||
| Wheat flour | SLE | 15 | 40 | 0.06 | The obvious differences of SERS signal between DON and ZEA, OTA, AFB1, T‐2, FB1 | [150] | ||||
| FL aptasensor | Corn flour | SLE | 30 | 45 | 1.87 | The restored FL intensity of DON showed a significantly higher value compared to AFB1, OTA, T‐2, and ZEA | [151] | |||
| Wheat flour | SLE | 15 | 40 | 0.08 | The obvious differences of FL signals between DON and ZEA, OTA, AFB1, T‐2, FB1 | [150] | ||||
| EC aptasensor | Corn flour | SLE | 45 | 90 | 6.9 × 10−6 | The obvious differences of current between DON and ZEA, T‐2, AFB1, FB1 | [152] | |||
| ZEA | LC‐MS/MS | Peanut | SLE | 120 | 30 | 5 | [105] | |||
| Pistachio | SLE | 120 | 30 | 10 | [105] | |||||
| Corn silage | QuEChERS | 13 | 44 | 9 | [135] | |||||
| Wheat | SLE | 120 | 30 | 5 | [105] | |||||
| Wheat | SLE | 100 | 15 | 1 | [106] | |||||
| Corn | SLE | 120 | 30 | 10 | [105] | |||||
| Corn | SLE | 100 | 15 | 0.5 | [106] | |||||
| Raisin | SLE | 120 | 30 | 2 | [105] | |||||
| LC‐FLD | Corn | ASE | 13 | 15 | 6 | [153] | ||||
| Wheat | ASE | 13 | 15 | 6 | [153] | |||||
| Rice | ASE | 13 | 15 | 5 | [153] | |||||
| Barley | ASE | 13 | 15 | 3 | [153] | |||||
| UPLC‐MS/MS | Alpinia oxyphylla | SLE | 35 | 32 | 0.3 | [114] | ||||
| Corn | QuEChERS | 60 | 25 | 2.5 | [115] | |||||
| HPLC‐MS/MS | Wheat flour | SLE | >65 | 30 | 1.6 | [107] | ||||
| Wheat | SPE | 90 | 24.2 | 2 | [134] | |||||
| Wheat flour | QuEChERS | 65.5 | 17 | 17.9 | [154] | |||||
| Corn | SPE | >11 | 25 | 0.22 | [109] | |||||
| HPLC‐FLD | Wheat | IAC | 1.12 | [137] | ||||||
| Wheat, corn flakes, bread | SLE | 23 | 20 | 2 | [155] | |||||
| Corn | IAC | 1.06 | [137] | |||||||
| Rice, wheat, oat, barley, corn | IAC | 0.5 | No interference from foreign peaks was observed at the retention times of AFB1, AFB2, AFG1, AFG2, OTA, and ZEA for the analytes | [156] | ||||||
| ELISA | Corn, corn noodles, corn cookies | SLE | 10 | 860 | 0.1 | 4.1%, 189.1%, and 43.9% CRs with α‐ZAL, β‐ZAL, and β‐ZEL | [157] | |||
| Corn | SLE | >8 | 195 | 0.13 | The negligible CRs with AFB1, DON, OTA, and T‐2 | [158] | ||||
| Rice, barley, corn | SLE | >30 | 140 | 0.15 | 121.5%, 65.3%, 21.5%, and 18.9% CRs with α‐ZAL, β‐ZAL, α‐ZEL, and β‐ZEL | [159] | ||||
|
Soybean meal, silage, sorghum, corn, distillers dried grains with soluble, total mixed ration |
SLE | 45 | 300 | 0.06 | The CRs of less than 11% and less than 1% with zearalanone and ZAL | [160] | ||||
| LFI | Corn | SLE | 30 | 11 | 3.6 | The negligible CR with CIT, OTA, DON, FB1, and AFB1 | [161] | |||
| Corn, wheat | SLE | 7 | 5 | 6 |
No CRs for multianalysis of DON and ZEA |
[142] | ||||
|
Soybean meal, silage, sorghum, corn, distillers dried grains with soluble, total mixed ration |
SLE | 45 | 5 | 10 | The CRs of less than 11% and less than 1% with zearalanone and ZAL | [160] | ||||
| SPR immunosensor | Wheat | SLE | 45 | 20 | 7.07 | 15.3% and 11.5% CRs with α‐ZEL and β‐ZEL | [144] | |||
| OWLS immunosensor | Corn | SLE | 20 | 2 × 10−6 | 25.2%,12.8%, and 2.7% CRs with α‐ZEL, α‐ZAL, and β‐ZAL | [162] | ||||
| EC immunosensor | Standard substance | 30 | 1.9 × 10−3 | Less than 2.4% CRs with both DON and T‐2. | [163] | |||||
|
MIP‐SPR biosensor |
Corn | 40 | 0.3 | 15%, 21%, 25%, and 27% selectivity efficiencies with α‐ZEL, β‐ZEL, α‐ZAL, zearalanone and α‐ZAL | [164] | |||||
|
MIP‐ FL biosensor |
Corn | 5 | 35%, 3%, and 4% CRs with ZOL, OTA, and AFB1. | [165] | ||||||
|
MIP‐ EC biosensor |
Corn | SLE | 5 | 15 | 0.2 | 10%, 9%, 7%, 10%, and 14% CRs with NIV, OTA, FB1, FB2, and DON. | [166] | |||
| EC aptasensor | Beers | 1.7 × 10−4 | No obvious change of current with AFT, α‐ZAL, β‐ZAL, β‐ZEL, and OTA | [167] | ||||||
| SERS aptasensor | Corn | SLE | 20 | 210 | 6.4 × 10−3 | The negligible Raman signal intensities with AFB1, OTA, DON, and FB1. | [168] | |||
| FL aptasensor | Corn | SLE | 30 | 150 | 0.126 | The negligible fluorescent‐signal changes with α‐ZEL, β‐ZEL, ZEA‐4‐G, ZEA‐4‐S, AFB1, AFB2, OTA, FB1, and FB2 | [169] | |||
| Beer | 150 | 0.007 | The negligible fluorescent‐signal changes with α‐ZEL, β‐ZEL, ZEA‐4‐G, ZEA‐4‐S, AFB1, AFB2, OTA, FB1, and FB2 | [169] | ||||||
3ADON, 3‐acetyldeoxynivalenol; 15ADON; 15‐acetyldeoxynivalenol; AFB: aflatoxin B; AFG, Aflatoxin G; AFT, aflatoxin; ASE, accelerated solvent extraction; CR, cross‐reactivity; CS, casein; DA, dopamine; EC:electrochemical; ECL, electrochemiluminescence; ELISA: enzyme‐linked immunosorbent assay; FL: fluorescence; FLD, fluorescence detection; GC, gas chromatography; HPLC, high‐performance liquid chromatography; FUS‐X: fusarenon X; Glu, glucose; IAC: immunoaffinity column; L‐cys, L‐cystein; L‐Hcys, L‐homocysteine; LAC, lactose; LC, liquid chromatography; LFI: lateral flow immunoassay; LLE: liquid–liquid extraction; MIP: molecularly imprinted polymer; MS, mass spectrometers; MS/MS, tandem mass spectrometry; NE, norepinephrine; OTA, ochratoxin A; OTB, ochratoxin B; OWLS, optical waveguide light‐mode spectroscopy; PAT, patulin; PDA,photodiode array; PEC, photoelectrochemical; QuEChERS: quick, easy, cheap, effective, rugged, and safe; SERS: surface‐enhanced Raman spectroscopy; SLE: solid–liquid extraction; SPE, solid phase extraction; SPM, sample preparation methods; SPR: surface plasmon resonance; UPLC, ultra‐performance liquid chromatography; ZEA‐4‐S, zearalenone‐4‐sulfate.
Extraction and precleaning methods
Primary extraction is essential for the determination of mycotoxins in various sample types. The cleanup step can eliminate interference from the extract and concentrate the analyte, and it is essential for the analysis of mycotoxins, especially at trace levels 102 . Currently, common approaches include solid phase extraction (SPE), multifunctional cleanup columns, liquid–liquid extraction (LLE), solid–liquid extraction (SLE), and immunoaffinity column (IAC) 170 . The selection of a method for mycotoxin extraction depends on the types of analytes. However, some methods can incur high costs, intricate procedures, and/or substantial time and solvent consumption. To minimize the sample treatment but prevent exposure to matrix effects, the “quick, easy, cheap, effective, rugged, and safe” (QuEChERS) sample preparation approach is a viable alternative. The QuEChERS method has been used for the extraction of mycotoxins from food samples, including dried fruits and cereals, as well as liquid samples, such as wine and beer 171 .
Chromatographic methods
There are many kinds of chromatographic analytical methods for mycotoxin analysis, such as thin‐layer chromatography (TLC), high‐performance TLC (HPTLC), gas chromatography (GC), high‐performance liquid chromatography (HPLC), and ultra‐performance liquid chromatography (UPLC). HPLC has emerged as the most widespread technique for mycotoxin analysis. By coupling to detectors, such as mass spectrometry (MS), ultraviolet (UV) detectors, visible detectors, and fluorescence (FL) detectors (FLDs), the compounds separated by chromatography can be further identified 20 . Currently, LC‒MS and liquid chromatography FL detection (LC‒FLD) are widely recognized as the standard methods for detecting Fusarium mycotoxins. There is no doubt that FLD is the most sensitive among all LC detectors. The sensitivity, precision, and accuracy of LC‒MS may vary depending on the mycotoxins, matrix, ionization technique, and sensitivity of the process. Due to ion suppression and matrix effects, LC‒MS often causes undesirable results for the quantitative measurement of mycotoxins. Tandem mass spectrometry (MS/MS) is the preferred detection method over FLD due to its ability to identify a wide range of both fluorescent and nonfluorescent mycotoxins, making it a cost‐effective choice 103 . With the ongoing development of technology, high‐throughput determination methods of single or multiple matrices using LC‒MS/MS have been reported. Steiner et al. 172 developed a pioneering multiclass quantitative method for the analysis of over 1200 biotoxins, pesticides, and veterinary drugs in complex feeds by LC–MS/MS.
In the past decade, liquid chromatography coupled with high‐resolution mass spectrometry (LC–HRMS) has turned from a research‐only technique into a costly tool for routine testing and high‐throughput food analysis in laboratories. Although these methods were initially developed for pesticide detection, mycotoxins are now the primary focus of LC–HRMS method development 173 . High resolution, in combination with the fast generation of product ion spectra, has the potential to minimize indistinct outcomes and streamline peak detection, but there remains a disparity between LC–HRMS and LC–MS/MS regarding the limits of detection (LOD) and of quantitation for most analytes. To address the gap for most analytes and enhance the applicability of mycotoxin trace analysis, various strategies for analyte enrichment, including SPE, have been incorporated into experimental protocols. However, these additional procedures invariably lead to a substantial augmentation in manual laboratory tasks and expenses, thereby diminishing the potential advantages of HRMS systems 173 , 174 . Mateus et al. 175 developed a UHPLC–HRMS multianalyte method for pistachio nuts. They evaluated different approaches to dispersive SPE for high‐lipid matrices; eventually, two procedures were validated. One involved the addition of enhanced matrix removal‐lipid for the detection of FUMs, and the other used a zirconium‐based material to achieve a slightly heightened sensitivity in analyzing G‐type AFs without including FUMs.
HPLC, UHPLC, and GC coupled with non‐MS detectors are also reference methods for Fusarium mycotoxin analysis. The current trend for the chromatographic analysis of Fusarium mycotoxins with non‐MS detection involves the advancement of multitoxin analysis with FLD, UV detection, photodiode array (PDA) detection, and so forth. Pi et al. 176 developed a novel method for the simultaneous determination of nine mycotoxins based on ultrasonic‐assisted aqueous two‐phase extraction coupled with solidifying organic drop‐dispersible liquid–liquid microextraction by HPLC with a diode array detector and FLD in series. The methodology effectively identified various mycotoxins in multiple foods. Furthermore, it can be utilized to effectively perform regular and extensive analyses of numerous mycotoxins within various samples. Lee et al. 177 built a simple and reliable HPLC‐UV method for the simultaneous determination of DON, NIV, DON‐3G, and NIV‐3G. This method involves a straightforward sample extraction with IAC purification and was successfully applied to analyze 31 different baby formulas and Korean rice wines available on the Korean market.
Immunological methods
Immunological detection methods rely on the antibody–antigen (Ab–Ag) binding relationship and vary from enzyme‐linked immunosorbent assay (ELISA) and lateral flow immunoassay (LFI) to advanced immunosensors. ELISA is a technique utilized to detect the presence and quantity of Ag binding in biological samples based on the principle of Ag–Ab interactions. In recent decades, numerous ELISA kits have been effectively commercialized for the detection of Fusarium mycotoxins. ELISA tests are portable, simple, fast, and do not require expensive analytical equipment. This continues to make ELISA tests popular. Nevertheless, ELISAs often show limited precision at low concentrations, and structurally similar mycotoxins or matrices can impede conjugate and Ab binding, causing errors in quantifiable mycotoxin ELISA measurements 103 .
LFI is a simple one‐step immunochromatographic paper assay that does not require complex instruments. It can be classified into two modes, competitive and sandwich; typically, LFI for mycotoxin detection adopts the competitive type. The basic LFI equipment consists of sample coating pads, conjugate‐release pads, absorbent pads, and membranes (also called detection pads) 178 . For the user, LFI is strikingly simple: after simply adding the sample onto a single paper lateral flow strip and a short incubation time, the qualitative or semiquantitative result of the test is revealed by the appearance of a test line, and quantification can then be conducted using an optical reader 179 . The exceptional advantages of LFI in terms of convenience, affordability, and rapidity make it particularly suitable for on‐site monitoring and rapid testing for Fusarium mycotoxin contamination in foods. In recent years, the advancement of novel nanomaterials has broadened the types of labels available for LFI of Fusarium mycotoxins. Colored nanoparticle (NP)‐based LFI is simpler and more convenient, and it shows significant potential for on‐site detection 180 . Due to the intricate nature of mycotoxin co‐occurrence, there is a growing need for simultaneous detection of multiple mycotoxins. To date, several multiplex LFIs for mycotoxins with excellent performance have been effectively developed. For example, Liu et al. 181 devised an innovative LFI integrated with gold nanoparticles (AuNPs) and time‐resolved FL microspheres. Their LFI is a smartphone‐based quantitative dual detection for multiplex mycotoxins in cereals, such as aflatoxin B1 (AFB1), ZEA, DON, T‐2, and FB1.
Biosensors
Biosensors are bioanalytical devices that incorporate biological recognition elements to bind target molecules and a signal transducer for converting the biorecognition event into a quantifiable signal. Biorecognition elements, including Abs, Ags, nucleic acids, and enzymes, are used for the identification and detection of target analytes. Based on a variety of bioinspired recognition elements, biosensors can be classified into immunosensors, aptasensors, and molecularly imprinted polymer (MIP)‐based sensors 179 . According to the principle of signal transduction, biosensors can be divided into electrochemical (EC) biosensors, optical biosensors, mass‐sensitive biosensors, thermal biosensors, and so on 182 . Compared to the other analytical methods mentioned above, biosensors offer a much simpler and more efficient means of dynamically monitoring reaction changes in real‐time with digital outputs. Biosensors not only reduce detection time but also enhance sensitivity, simplicity, robustness, and reusability, enabling the development of cost‐effective, high‐throughput screening methods for mycotoxins 179 .
Immunosensors, as novel and widely utilized analytical instruments, use Ab as the recognition element and a transducer to convert the Ag–Ab binding event into a quantifiable physical signal. Due to the superior specificity of the Ag–Ab immunoreaction, immunosensors show superior selectivity and sensitivity 183 . Various immunosensors have been developed based on the different mechanisms of signal variations, including FL, colorimetric, chemiluminescence, electrochemiluminescence, surface plasmon resonance (SPR), surface‐enhanced Raman spectroscopy (SERS), and EC immunosensors. By combining the exceptional specificity of Abs and the remarkable sensitivity of FL detection, FL immunosensors have recently emerged as highly favored contenders for mycotoxin detection 183 . With advancements and breakthroughs in nanotechnology, a diverse array of nanomaterial semiconductors, including semiconductor quantum dots (QDs), QD nanobeads (QBs), carbon dots (CDs), fluorescent metallic NPs, and upconversion nanoparticles (UCNPs), with unique photostability, bright FL, and good biocompatibility have garnered immense attention regarding their use in the construction of FL immunosensors 184 . Yang et al. 185 devised a novel FL immunosensor detection platform that integrates multicolor UCNP barcoding technology with smartphone‐based portable devices for simultaneous analysis of multiple mycotoxins (AFB1, ochratoxin A [OTA], and ZEA). The quantitative detection platform demonstrated feasibility and reliability, with a LOD of 1 ng that surpassed the values obtained from standard assays. SERS is a surface‐sensitive vibrational spectroscopy technique for the detection and characterization of analytes that are adsorbed on or close to the surface of plasmonic nanostructures. SERS integrates the advantages of the molecular specificity of Raman spectroscopy and the optical sensitivity of plasmonic nanostructures to boost the Raman signal, greatly extending the role and application field of standard Raman spectroscopy 183 . SERS significantly amplifies the Raman signal, thereby expanding the scope and applicability of conventional Raman spectroscopy in a profound manner. SERS immunosensors, which integrate the SERS labeling technique with Ag–Ab specific interactions, have emerged as novel immunosensing devices for mycotoxins. Li et al. 186 developed a SERS immunosensor to simultaneously detect AFB1, ZEA, and OTA by using AuNPs labeled with 5,5‐dithiobis(succinimidyl‐2‐nitrobenzoate) as a Raman reporter. The SERS immunosensor determination method demonstrated results that are consistent with conventional instrumental analysis.
Aptamers are novel recognition elements with exceptional affinity and specificity that have the potential to serve as recognition molecules in aptasensors (aptamer‐based biosensors) for the efficient and swift identification of various targets 187 . Aptamers can be divided into two categories, DNA/RNA‐based aptamers and peptide aptamers. Currently, aptamers for the detection of mycotoxins are a class of single‐stranded DNAs or RNAs that undergo screening through the systematic evolution of ligands by exponential enrichment, which can selectively bind to different ligands through noncovalent bonds 188 . The functional similarity of aptamers to Abs makes them widely applicable in the field of biosensors. Compared to Abs, aptamers are low cost, have a longer shelf life and good stability, even at elevated temperatures, and are easy to modify and synthesize 189 . As with immunosensors, according to the signal variation mechanism, aptasensors can also be classified as FL aptasensors, EC aptasensors, SPR aptasensors, SERS aptasensors, CEL aptasensors, and so on. EC aptasensors use electrodes as sensing units and electrochemical workstations as signal transformation systems 190 . Aptamers are typically immobilized on the surface of the electrode; the specific molecular interactions between molecules on the electrode surface result in the conversion of target binding into electrical signals. These electrical signals, such as current, resistance, potential, or capacitance, are transmitted to a computer for quantitative or qualitative analysis of the target 187 . Zhang et al. 191 devised an electrochemical aptasensor‐based target‐induced strand displacement strategy to achieve highly sensitive detection of T‐2. The aptasensor was highly specific, stable, and suitable for T‐2 detection in real samples. SPR is a phenomenon in which the electrons in a metal surface layer are excited by photons of incident light with a certain angle of incidence and then propagate parallel to the metal surface. With a constant light source wavelength and a thin metal surface, the angle that triggers SPR depends on the refractive index of the material near the metal surface. The affinity binding interaction on the surface of thin metal films can cause a small change in the reflective index of the sensing medium, which can hinder the occurrence of SPR. SPR aptasensors can detect those changes on the optical transducer surface, and optical transduction can directly convert the molecular binding event into a physically measurable signal, which is proportional to the concentration of analyte molecules 192 . As a label‐free analytical strategy, SPR aptasensors have the capability to detect multiple mycotoxins simultaneously with real‐time monitoring, high sensitivity, good specificity, minimal sample preparation requirements, and high‐throughputdetection 192 . Wei et al. 144 effectively and simultaneously detected OTA, DON, AFB1, and ZEA in wheat and corn using SPR aptasensors that demonstrated high sensitivity, good linearity, and specificity.
MIPs, also referred to as “plastic antibodies,” are bespoke synthetic receptors that use the molecular imprinting technique. Following the general molecular imprinting method, imprint molecules (templates) are added along with functional monomers and cross‐linkers, which are polymerized under appropriate conditions. After copolymerization, the templates are extracted, leaving the 3D matrix with cavities that possess a matching shape, size, and chemical functionality with the template. The resulting cavities possess the ability to selectively reassociate with the template upon subsequent exposure and become synthetic receptors 174 , 193 . The interactions between the polymers and the templates are similar to those between Abs and Ags. MIPs provide a cost‐effective and easily prepared approach for targeted template recognition with outstanding biocompatibility, repeatability, and broad utility. MIPs are particularly robust in extreme environments, such as elevated pressures, fluctuations in pH levels, and exceptionally high or low temperatures. By integrating MIPs with various sensing reporter systems (electrical, EC, optical methods, etc.), a valuable device can be developed for monitoring or screening purposes 194 . Utilizing a blend of MIP membranes (ZEA‐selective urethane acrylate MIP membranes) and the Spotxel® Reader smartphone application (Sycasys Software GmbH), a miniature sensor was used to analyze the natural FL of ZEA in cereals in the field 195 . NPs show a high level of sensitivity, yet their selectivity is limited. Therefore, the combination of MIPs and NPs can result in MIP composites with high sensitivity and selectivity. To date, several NPs, including QDs, UCNPs, carbon NPs (CNPs), AuNPs, magnetic NPs, and metal‐organic frameworks, have played important roles in mycotoxin analysis with MIPs 193 . The molecularly imprinted polymer nanoparticle‐based assay (MINA) was developed for the determination of FB1—the molecularly imprinted nanoparticles replace the primary Ab used in a competitive ELISA. The MINA showed a high level of specificity and did not show any cross‐reactivity with other mycotoxins, including AFB1, CIT, DON, ZEA, and FB2. The results of the MINA agreed with those obtained using traditional ELISA and HPLC methods 196 . Calahorra‐Rio et al. 197 developed a new molecularly imprinted magnetic nanobead that can specifically extract ZEA from river and tap water for further analysis using HPLC–FLD. Their findings indicate that the magnetic nanobead showed exceptionally accurate and consistent ZEA detection in real liquid samples.
THE MANAGEMENT STRATEGY FOR FUSARIUM MYCOTOXIN CONTAMINATION
Fusarium mycotoxins can be produced by several fungi in several stages of several crops, so a single management strategy cannot fully control Fusarium mycotoxin contamination. However, a series of practices, including agronomic, chemical, physical, and biological methods, can be implemented to avoid the spread of mycotoxins and to minimize their frequency in food products during preharvest and postharvest stages (Figure 7).
Figure 7.
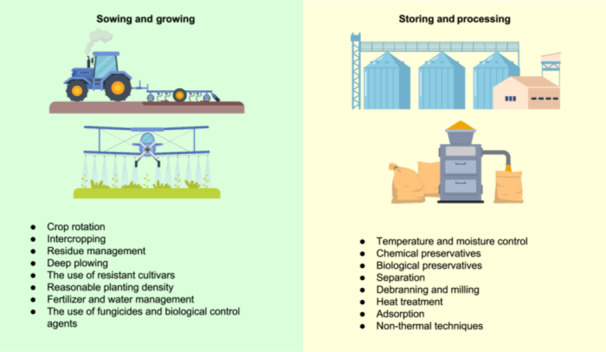
Proper practices to minimize Fusarium mycotoxin contamination. There are a series of management strategies that can be utilized to control Fusarium mycotoxins during the preharvest (sowing and growing) and postharvest stages (storing and processing). During the preharvest stage, these strategies place emphasis on preventing infection by Fusarium fungi. During the postharvest stage, management strategies are more focused on controlling the production of new mycotoxins and the detoxification and removal of existing mycotoxins.
Agronomic method
Fusarium fungi can survive in plant residues and on wild grasses, existing as mycelia, conidia, or perithecia. Therefore, residue management, tilling, and deep plowing can reduce the primary Fusarium inoculum that causes infections and the presence of mycotoxins 12 . Reduced or no tillage practices can contribute to increased DON contamination of wheat and maize 198 . Proper crop rotation can substantially mitigate Fusarium disease and mycotoxin contamination. An unfavorable practice in crop rotation is the consecutive cultivation of cereal plants 12 . Qiu et al. 199 collected 90 wheat samples in China during 2013 and 2014, revealing that DON contamination was more prevalent in the rice–wheat rotation, while ZEA accumulation was found to be higher in the maize–wheat rotation. Crop rotation with noncereals can be a more favorable method for limiting Fusarium mycotoxin contamination; legumes, brassicas, and root crops might be better forecrops 12 . Moreover, the implementation of intercropping is a good method for controlling Fusarium mycotoxin contamination. Drakopoulos et al. 200 found that the use of white mustard or Indian mustard as an intercrop with maize can reduce DON in winter wheat compared to maize grown as a sole crop in maize–wheat rotation fields under reduced tillage. The cultivation system, conventional or organic, also affects Fusarium mycotoxin contamination of cereals. Bernhoft et al. 201 conducted a comprehensive analysis of available published studies (1991–2017) of controlled field experiments, comparing DON, ZEA, and T‐2/HT‐2 in grains from organic and conventional cultivation systems. Summary of the results revealed that organic production showed lower mycotoxin levels in 24 cases, no significant difference in 16 cases, and higher levels in only two cases.
Host resistance
The most economical and sustainable approach to controlling Fusarium disease and mycotoxin contamination is through the cultivation of varieties that show resistance or partial resistance to Fusarium species, whether because of traditional breeding or through use of transgenic technology. Taking the case of FHB in wheat, there are five types of resistance: Type I (resistance to initial infection), Type II (resistance to disease spread within infected heads), Type III (resistance to mycotoxin accumulation), Type IV (resistance to kernel damage), and Type V (tolerance) 202 . These FHB resistance traits of wheat are governed by multiple quantitative trait loci (QTLs). Over 250 QTLs for FHB have been found on all 21 chromosomes in various wheat genotypes 203 . Fhb1 on chromosome 3BS is the most important QTL. The Chinese wheat cultivar Sumai 3, known for carrying the Fhb1 gene, is widely acknowledged as the superior source of FHB resistance and has been extensively utilized as a parent in numerous breeding programs 204 . Transgenic germplasms can offer novel supplementary resources for FHB management. Wheat resistance to F. graminearum could be significantly enhanced by overexpression of genes that encode defense signaling pathway‐related proteins, cell wall‐degrading enzyme inhibitors, and detoxification proteins 22 . McLaughlin et al. 205 showed that the overexpression of AtLTP4.4, a nonspecific lipid transfer protein‐encoding gene of Arabidopsis thaliana, in transgenic wheat, can significantly reduce F. graminearum infection and DON contamination in the field. Based on RNA silencing, plants naturally have a defense system to stave off viral invasion. This feature has been harnessed to advance host‐induced gene silencing (HIGS) technology to control other plant pathogens by silencing pathogen genes in plants during infection. HIGS is also an effective control strategy against FHB in wheat. Recently, many researchers have successfully silenced important genes encoding cytochrome P450, chitin synthase, protein kinases, and others in F. graminearum, Fusarium oxysporum, F. culmorum, and F. verticillioides, resulting in reduced mycotoxin production 206 .
Chemical method
Currently, due to the absence of efficacious disease‐resistant cultivars, the utilization of chemical fungicides remains the primary strategy for Fusarium disease management. There are several common types of fungicides for controlling Fusarium diseases, including benzimidazoles (benomyl, carbendazim, thiophanate‐methyl, and thiabendazole), triazoles (triadimefon, tebuconazole, diniconazole, and propiconazole), and strobilurins (azoxystrobin and pyraclostrobin). Strobilurin treatment can induce DON formation, so strobilurins are unlikely to be effective 207 . Triazoles, a group of demethylation inhibitor fungicides, are the gold standard for controlling FHB in wheat worldwide, while carbendazim is the most widely used fungicide in China for this purpose 207 , 208 . In addition, phenamacril, a myosin I inhibitor and cyanoacrylate fungicide, is an efficacious and highly species‐specific fungicide for FHB in China. Phenamacril is not only capable of inhibiting the mycelial growth of a few Fusarium species, including F. graminearum, Fusarium asiaticum, F. verticillioides, and F. oxysporum, but also significantly disrupts DON‐toxisome formation to hinder DON biosynthesis 209 . Although spraying fungicides is a very effective method, the use of chemical fungicides comes with its own set of challenges. In addition to the well‐known environmental problems caused by fungicides, fungicide‐resistant population of Fusarium species are also an important concern. Due to prolonged and intensive use of fungicides, the benzimidazole‐ and triazole‐resistant strains of F. graminearum have become feared throughout world agriculture. There was a high frequency of carbendazim‐resistant F. graminearum in China. In response, the local government of Jiangsu Province, where large carbendazim‐resistant F. graminearum population had appeared, even proposed a fallow subsidy policy 210 . Regarding triazoles, tebuconazole‐resistant F. graminearum strains have been found in Argentina, China, Germany, and the United States 207 . Therefore, the development of new fungicides for the management of Fusarium diseases and mycotoxin contamination is urgently needed.
Chemical preservatives can be added to stored grain, particularly in the case of damp grains intended for animal feed. Common chemical preservatives include mixtures of aliphatic acid salts and antioxidants. The former has demonstrated efficacy in controlling FB1 production of F. proliferatum on irradiated maize kernels 211 . The two antioxidants, butylated hydroxyanisol and propyl paraben, were shown to reduce both the growth and FUM production of F. verticillioides and F. proliferatum in culture media and maize grain 212 . Recently, considering the adverse effects of synthetic preservatives, there has been a growing trend toward utilizing plant essential oils (EOs) as viable alternatives for mycotoxin management.
Biological control method
Biological control is an eco‐friendly approach that uses living organisms or their derivatives to control pests. The useful biological control agents (BCAs) for Fusarium diseases and mycotoxin contamination include beneficial bacteria, fungi, actinomycetes, and mycoviruses. BCAs can protect crops against pathogenic Fusarium fungi before harvest, thereby effectively mitigating the risk of mycotoxin contamination in the food chain. The mechanisms of BCAs encompass competition for space and nutrients, mycoparasitism, synthesis of antifungal metabolites, cross‐protection, promotion of plant growth, and induction of host plant resistance 213 .
Among the bacterial BCAs, two genera that secrete antifungal compounds, Bacillus (such as Bacillus velezensis, Bacillus megaterium, and Bacillus subtilis) and Pseudomonas (including Pseudomonas aeruginosa, Pseudomonas frederiksbergensis, Pseudomonas fluorescens, and Pseudomonas simiae), are the most widely used and can effectively reduce Fusarium infection and mycotoxin contamination under field conditions 214 . Bacillus contain three important antifungal lipopeptides that inhibit Fusarium pathogens, including Bacillomycin D, surfactin, and fengycin (synonymous with plipastatin) 215 . Bacillomycin D and fengycin can inhibit the growth of hyphae, cause morphological alterations or destruction of cell walls and plasma membranes, and trigger the cell lysis of F. graminearum and Fusarium moniliforme through the accumulation of ROS. Moreover, Iturin A, a kind of Bacillomycin D, can also control the T‐2 toxin synthesis of F. oxysporum by inhibiting TRI5 expression 216 . Regarding F. moniliforme, surfactin can inhibit hyphal growth and induce ROS, damaging DNA and protein in living cells 217 . In addition to direct interactions, bacterial BCAs can also protect plants against Fusarium pathogens in indirect ways. Chen et al. 218 suggested that B. velezensis LM2303 has various biocontrol mechanisms against F. graminearum, such as enhancing wheat systemic resistance, facilitating wheat plant growth, and outcompeting for space and nutrient competition via efficient colonization and antibacterial metabolites.
The genera Aureobasidium, Cladosporium, Clonostachys, Cryptococcus, Sarocladium, and Trichoderma are the representative fungal BCAs for Fusarium disease management. Trichoderma is a notable example of a fungal antagonist, as it not only inhibits the growth and reproduction of Fusarium fungi through mycoparasitism, antibiotics, promotion of plant growth, and the induction of defense responses in host plants, but also has the ability to control the biosynthesis of Fusarium mycotoxins and directly degrade Fusarium mycotoxins 219 . Błaszczyk et al. 220 reported that Trichoderma atroviride shows significant inhibitory effects on the biosynthesis of mycotoxins (DON, 3ADON, 15ADON, NIV, ZEA, BEA, and MON) of Fusarium avenaceum, Fusarium cerealis, F. culmorum, F. graminearum, and Fusarium temperatum. Tian et al. 221 , 222 indicated that Trichoderma fungi can convert ZEA into ZEA sulfate and ZEL sulfate by sulfation and glycosylate TRIs into glycosylated forms. Galletti et al. 223 found that Trichoderma gamsii B21 can remove FUMs from liquid cultures, but the mechanisms were unclear. Recently, the utilization of endophytic fungi as BCAs has become regarded as a compelling strategy for controlling plant disease. Kemp et al. 224 found that the wheat endophytic fungus Sphingobacterium zeae NRRL 34560 can induce defense responses in wheat, effectively controlling FHB and DON contamination in a greenhouse.
Mycoviruses, also known as fungal viruses, are parasitic viruses that infect a wide range of filamentous fungi and yeasts, including Fusarium fungi. Mycoviruses typically do not exert phenotypic effects on their hosts; however, some can induce negative consequences, such as hypovirulence 225 . These hypovirulence‐related mycoviruses have great potential as BCAs. In total, mycoviruses have been documented in 13 Fusarium species, including F. asiaticum, Fusarium boothii, Fusarium circinatum, Fusarium coeruleum, Fusarium globosum, F. graminearum, F. incarnatum, Fusarium langsethiae, F. oxysporum, Fusarium poae, Fusarium pseudograminearum, Fusarium solani, and Fusarium virguliforme. Most Fusarium mycoviruses establish latent infections, but some, including Fusarium graminearum virus 1 (FgV1), Fusarium graminearum virus‐ch9, Fusarium graminearum hypovirus 2 (FgHV2), Fusarium oxysporum f. sp. dianthi mycovirus 1, and Fusarium pseudograminearum megabirnavirus 1 (FpgMBV1), cause hypovirulence 225 . Moreover, FgV1 and FgHV2 infections can reduce DON production in F. graminearum 226 , 227 . Li et al. 228 found that FpgMBV1 eliminates DON accumulation by downregulating the expression of TRI biosynthetic genes. However, most research in this area is still in the laboratory stage and has not been applied in the field.
After harvest, beneficial microorganisms can be used to control Fusarium mycotoxin contamination. Lactic acid bacteria (LAB), which are frequently used as biological preservatives, not only can prevent Fusarium fungal growth but can also control mycotoxin contamination in food and feed. Strains of Lactobacillus, Bifidobacterium, Lactococcus, Leuconostoc, Pediococcus, Propionibacterium, and Streptococcus have been used to control mycotoxin contamination 229 . LAB produce many antifungal metabolites, such as organic acids, hydrogen peroxide, diacetyl, rutherin, reutericyclin, acetoin, bacteriocins, and bacteriocin‐like inhibitory substances 230 . In addition to its antifungal activity, LAB possess the ability to adsorb, degrade, or detoxify Fusarium mycotoxins. On one hand, LAB can assimilate mycotoxins toward their cell wall operative groups; on the other hand, LAB can degrade mycotoxins via metabolic apparatus or enzymes 229 .
A drop in the utilization of synthetic preservation has occurred due to its side effects, such as resistance development in pests, nonbiodegradable characteristics, and toxic effects on nontargeted organisms. Therefore, as green preservatives, plant EOs have become widely used substitutes for synthetic preservatives. In the past decade, cinnamon, clove, eucalyptus, fennel, oregano, rosemary, palmarosa, and thyme were the most frequently used EOs to control mycotoxigenic fungi and their mycotoxins. EOs can prevent fungal infection and mycotoxin biosynthesis in many ways, such as the inhibition of fungal growth, destruction of cell permeability, disruption of the electron transport chain, and modulation of gene expression patterns and metabolic processes 231 . Due to the development of nanotechnology, nanoencapsulation of EOs has emerged as a novel strategy to increase the stability of EO constituents, extend their applicability, and overcome their major limitations by controlled release. Singh et al. 232 fabricated clove oil nanoemulsions and found that they can control the growth of F. proliferatum and reduce the FB1 and FB2 contents of maize.
Physical method
Physical methods can be used to eliminate mycotoxins during postharvest storage and processing; these include separation, debranning and milling, heat treatment, radiation, and adsorption. Mycotoxins are mainly found in the moldy, fragmented, and discolored parts of grains, and the specific gravity of mycotoxin‐contaminated cereals is lower than that of uncontaminated cereals 233 . These characteristics enable Fusarium‐damaged grains to be segregated by aspiration, image processing techniques, gravity separation, and photoelectric separation. In general, the external layer of grains tends to show higher levels of mycotoxin contamination. Partial debranning of 10% of the wheat grain weight can result in a 64% reduction in DON content 234 . Therefore, debranning and milling procedures are important for the decontamination of Fusarium mycotoxins. After polishing, the Fusarium mycotoxin content of white rice can be markedly decreased 235 . Regarding the milling process, Fusarium mycotoxins tend to accumulate in the outer fractions of wheat grains (bran, flour shorts, screenings, and middlings), which are primarily utilized as animal feeds, and lower concentrations are found in the inner fractions (flour or semolina) intended for human consumption 236 .
There are a series of innovative physical methods to control Fusarium mycotoxin contamination that are usually based on nonthermal techniques: cold atmospheric plasma (CAP), electron beam irradiation, pulsed light, and so on. The CAP process makes use of an ionized gas at near room temperature to inactivate microorganisms 237 . CAP is also successful at degrading mycotoxins due to the oxidizing potential of plasma. Compared to conventional and other nonthermal approaches, CAP shows rapid efficacy in controlling fungi and mycotoxins, exerts minimal influence on product quality, and requires only low energy consumption 238 . Feizollahi et al. 239 found that CAP treatment can reduce DON in barley by 49% in 6 min, and Wielogorska et al. 240 reported a significant decrease (66%) in FB1 in maize following a 10‐min treatment with CAP. So, CAP technology represents remarkable progress in the food industry. Unfortunately, prior studies have only been successful on the laboratory scale. Therefore, additional research is warranted to demonstrate the effectiveness within wide‐scale food production and different food matrices, as well as of different types of gasses 233 .
CONCLUDING REMARKS
In this review, we elucidate the effects of Fusarium mycotoxin contamination on food safety and summarize the countermeasures to prevent or mitigate harm to humans. In addition, we discuss how CC influences Fusarium mycotoxin contamination—many models have been constructed on historical or current datasets of climatic conditions to anticipate the interactions of Fusarium mycotoxins with CC. However, most models lack the [CO2] factor, which exerts a significant influence on the growth of both crops and fungi. The ultimate goal of controlling Fusarium mycotoxin contamination is to protect human health. Nevertheless, few studies have estimated the transfer of mycotoxins from fields to human foods or their eventual impact on human health under different CC scenarios.
Fusarium mycotoxins cannot be completely eliminated worldwide. All we can do is to reduce contamination to acceptable levels that do not threaten human health. Therefore, a number of detection technologies and management strategies have been developed. Both agricultural production and industrial food processing are economic activities; therefore, scholars and practitioners should consider both the economic benefits and the costs of the application of Fusarium mycotoxin countermeasures to producers. With the improvement of living standards, environmental stability begins to take precedence over production efficiency and economic benefits. The development and recommended use of farm chemicals is a representative example. So, the development of Fusarium mycotoxin countermeasures should be based on a balance among these three factors. Good agricultural practices (GAPs) are a set of guidelines for producing safe and healthy food and nonfood agricultural products through on‐farm and post‐production practices, with due consideration of the sustainability of economic, social, and environmental aspects 241 . With regard to the control of Fusarium mycotoxins, GAP is of great significance and has been applied in many countries 242 . Nevertheless, it is not easy to change the agricultural practices of smallholder farmers. These changes may require more communication, education, and support from local governments, especially in developing countries.
ACKNOWLEDGMENTS
This work was supported by the National Key Research and Development Program of China (2022YFD1901305) and the Agricultural Science and Technology Innovation Program (CAAS‐ZDRW202416). Thanks to Academician Peiwu Li for his guidance on this paper. We thank Dr. Ying Zhao (the associate professor of Henan Agricultural University), Dr. Binnian Tian (the associate professor of Southwest University), and Dr. Jie Wang (the assistant researcher of Peking University Institute of Advanced Agricultural Sciences) for photos in Figure 1.
Qu Z, Ren X, Du Z, Hou J, Li Y, Yao Y, et al. Fusarium mycotoxins: The major food contaminants. mLife. 2024;3:176–206. 10.1002/mlf2.12112
Editor: Lei Cai, Institute of Microbiology, Chinese Academy of Sciences, China
Contributor Information
Yanpo Yao, Email: yao7707@126.com.
Yi An, Email: anyi@caas.cn.
REFERENCES
- 1. Purvis A, Hector A. Getting the measure of biodiversity. Nature. 2000;405:212–219. [DOI] [PubMed] [Google Scholar]
- 2. Xu J. Assessing global fungal threats to humans. mLife. 2022;1:223–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pathakumari B, Liang G, Liu W. Immune defence to invasive fungal infections: a comprehensive review. Biomed Pharmacother. 2020;130:110550. [DOI] [PubMed] [Google Scholar]
- 4. Peng Y, Li SJ, Yan J, Tang Y, Cheng JP, Gao AJ, et al. Research progress on phytopathogenic fungi and their role as biocontrol agents. Front Microbiol. 2021;12:670135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pleadin J, Frece J, Markov K. Mycotoxins in food and feed. In: Toldra F, editor. Advances in food and nutrition research. Cambridge, UK: Elsevier; 2019. p. 297–345. [DOI] [PubMed] [Google Scholar]
- 6. El‐Sayed RA, Jebur AB, Kang W, El‐Demerdash FM. An overview on the major mycotoxins in food products: characteristics, toxicity, and analysis. J Future Foods. 2022;2:91–102. [Google Scholar]
- 7. Boutrif E, Canet C. Mycotoxin prevention and control: FAO programmes. Revue de Médecine Vétérinaire. 1998;149:681–694. [Google Scholar]
- 8. Eskola M, Kos G, Elliott CT, Hajšlová J, Mayar S, Krska R. Worldwide contamination of food‐crops with mycotoxins: validity of the widely cited ‘FAO estimate’ of 25%. Crit Rev Food Sci Nutr. 2019;60:2773–2789. [DOI] [PubMed] [Google Scholar]
- 9. Godswill Awuchi C, Nyakundi Ondari E, Josiah Eseoghene I, Twinomuhwezi H, Otuosorochi Amagwula I, Morya S. Fungal growth and mycotoxins production: types, toxicities, control strategies, and detoxification. In: Sultan S, Singh GKS, editors. Fungal reproduction and growth. London, UK: IntechOpen; 2022. p. 1–21. [Google Scholar]
- 10. Venkatesh N, Keller NP. Mycotoxins in conversation with bacteria and fungi. Front Microbiol. 2019;10:403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Munkvold GP, Proctor RH, Moretti A. Mycotoxin production in Fusarium according to contemporary species concepts. Annu Rev Phytopathol. 2021;59:373–402. [DOI] [PubMed] [Google Scholar]
- 12. Mielniczuk E, Skwaryło‐Bednarz B. Fusarium head blight, mycotoxins and strategies for their reduction. Agronomy. 2020;10:509. [Google Scholar]
- 13. Geiser DM, Al‐Hatmi AMS, Aoki T, Arie T, Balmas V, Barnes I, et al. Phylogenomic analysis of a 55.1‐kb 19‐gene dataset resolves a monophyletic Fusarium that includes the Fusarium solani species complex. Phytopathology®. 2021;111:1064–1079. [DOI] [PubMed] [Google Scholar]
- 14. Tan H, Zhou H, Guo T, Zhou Y, Zhang Q, Zhang Y, et al. Recent advances on formation, transformation, occurrence, and analytical strategy of modified mycotoxins in cereals and their products. Food Chem. 2023;405:134752. [Google Scholar]
- 15. Zhang Z, Nie D, Fan K, Yang J, Guo W, Meng J, et al. A systematic review of plant‐conjugated masked mycotoxins: occurrence, toxicology, and metabolism. Crit Rev Food Sci Nutr. 2020;60:1523–1537. [DOI] [PubMed] [Google Scholar]
- 16. O'Donnell K, Rooney AP, Proctor RH, Brown DW, McCormick SP, Ward TJ, et al. Phylogenetic analyses of RPB1 and RPB2 support a middle Cretaceous origin for a clade comprising all agriculturally and medically important fusaria. Fungal Genet Biol. 2013;52:20–31. [DOI] [PubMed] [Google Scholar]
- 17. McCormick SP, Stanley AM, Stover NA, Alexander NJ. Trichothecenes: from simple to complex mycotoxins. Toxins. 2011;3:802–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Haschek WM, Voss KA. Mycotoxins. In: Haschek Wm, Rousseaux Cg, Wallig Ma, Bolon B, Ochoa R, Mahler Bw editors. Haschek and Rousseaux's handbook of toxicologic pathology. London, UK: Elsevier; 2013. p. 1187–1258. [Google Scholar]
- 19. Desjardins AE, Proctor RH. Molecular biology of Fusarium mycotoxins. Int J Food Microbiol. 2007;119:47–50. [DOI] [PubMed] [Google Scholar]
- 20. Ji F, He D, Olaniran AO, Mokoena MP, Xu J, Shi J. Occurrence, toxicity, production and detection of Fusarium mycotoxin: a review. Food Prod Process Nutr. 2019;1:6. [Google Scholar]
- 21. Alexander NJ, Proctor RH, McCormick SP. Genes, gene clusters, and biosynthesis of trichothecenes and fumonisins in Fusarium . Toxin Rev. 2009;28:198–215. [Google Scholar]
- 22. Chen Y, Kistler HC, Ma Z. Fusarium graminearum trichothecene mycotoxins: biosynthesis, regulation, and management. Annu Rev Phytopathol. 2019;57:15–39. [DOI] [PubMed] [Google Scholar]
- 23. Nahle S, El Khoury A, Atoui A. Current status on the molecular biology of zearalenone: its biosynthesis and molecular detection of zearalenone producing Fusarium species. Eur J Plant Pathol. 2020;159:247–258. [Google Scholar]
- 24. Kim YT, Lee YR, Jin J, Han KH, Kim H, Kim JC, et al. Two different polyketide synthase genes are required for synthesis of zearalenone in Gibberella zeae . Mol Microbiol. 2005;58:1102–1113. [DOI] [PubMed] [Google Scholar]
- 25. Yao Y, Long M. The biological detoxification of deoxynivalenol: a review. Food Chem Toxicol. 2020;145:111649. [DOI] [PubMed] [Google Scholar]
- 26. EFSA Panel on Contaminants in the Food Chain (CONTAM), Knutsen HK, Alexander J, Barregard L, Bignami M, Bruschweiler B, et al. Risks to human and animal health related to the presence of deoxynivalenol and its acetylated and modified forms in food and feed. EFSA J. 2017;15:4718. [Google Scholar]
- 27. Yan P, Liu Z, Liu S, Yao L, Liu Y, Wu Y, et al. Natural occurrence of deoxynivalenol and its acetylated derivatives in Chinese maize and wheat collected in 2017. Toxins. 2020;12:200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hooft JM, Bureau DP. Deoxynivalenol: mechanisms of action and its effects on various terrestrial and aquatic species. Food Chem Toxicol. 2021;157:112616. [DOI] [PubMed] [Google Scholar]
- 29. Zhou H‐R, Jia Q, Pestka JJ. Ribotoxic stress response to the trichothecene deoxynivalenol in the macrophage involves the SRC family kinase Hck. Toxicol Sci. 2005;85:916–926. [DOI] [PubMed] [Google Scholar]
- 30. Yu YH, Lai YH, Hsiao FSH, Cheng YH. Effects of deoxynivalenol and mycotoxin adsorbent agents on mitogen‐activated protein kinase signaling pathways and inflammation‐associated gene expression in porcine intestinal epithelial cells. Toxins. 2021;13:301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liao S, Tang S, Tan B, Li J, Qi M, Cui Z, et al. Chloroquine improves deoxynivalenol‐induced inflammatory response and intestinal mucosal damage in piglets. Oxid Med Cell Longevity. 2020;2020:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. You L, Zhao Y, Kuca K, Wang X, Oleksak P, Chrienova Z, et al. Hypoxia, oxidative stress, and immune evasion: a trinity of the trichothecenes T‐2 toxin and deoxynivalenol (DON). Arch Toxicol. 2021;95:1899–1915. [DOI] [PubMed] [Google Scholar]
- 33. Biswas SK. Does the interdependence between oxidative stress and inflammation explain the antioxidant paradox? Oxid Med Cell Longevity. 2016;2016:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Starkov AA. The role of mitochondria in reactive oxygen species metabolism and signaling. Ann NY Acad Sci. 2008;1147:37–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Boice A, Bouchier‐Hayes L. Targeting apoptotic caspases in cancer. Biochim Biophys Acta. 2020;1867:118688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ding C, Shi X, Guan Y, Li X. Deoxynivalenol induces carp neutrophil apoptosis and necroptosis via CYP450s/ROS/PI3K/AKT pathway. Aquaculture. 2021;545:737182. [Google Scholar]
- 37. Yang J, Wang J, Guo W, Ling A, Luo A, Liu D, et al. Toxic effects and possible mechanisms of deoxynivalenol exposure on sperm and testicular damage in BALB/c mice. J Agricult Food Chem. 2019;67:2289–2295. [DOI] [PubMed] [Google Scholar]
- 38. Zhang Z, Fan K, Meng J, Nie D, Zhao Z, Han Z. Deoxynivalenol hijacks the pathway of Janus kinase 2/signal transducers and activators of transcription 3 (JAK2/STAT‐3) to drive caspase‐3‐mediated apoptosis in intestinal porcine epithelial cells. Sci Total Environ. 2023;864:161058. [DOI] [PubMed] [Google Scholar]
- 39. Gu X, Guo W, Zhao Y, Liu G, Wu J, Chang C. Deoxynivalenol‐induced cytotoxicity and apoptosis in IPEC‐J2 cells through the activation of autophagy by inhibiting PI3K‐AKT‐mTOR signaling pathway. ACS Omega. 2019;4:18478–18486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wang X, Liu Q, Ihsan A, Huang L, Dai M, Hao H, et al. JAK/STAT pathway plays a critical role in the proinflammatory gene expression and apoptosis of RAW264.7 cells induced by trichothecenes as DON and T‐2 toxin. Toxicol Sci. 2012;127:412–424. [DOI] [PubMed] [Google Scholar]
- 41. Chen J, Wen J, Tang Y, Shi J, Mu G, Yan R, et al. Research progress on fumonisin B1 contamination and toxicity: a review. Molecules. 2021;26:5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Victor P, Sarada D, Ramkumar KM. Crosstalk between endoplasmic reticulum stress and oxidative stress: focus on protein disulfide isomerase and endoplasmic reticulum oxidase 1. Eur J Pharmacol. 2021;892:173749. [DOI] [PubMed] [Google Scholar]
- 43. Hu H, Tian M, Ding C, Yu S. The C/EBP homologous protein (CHOP) transcription factor functions in endoplasmic reticulum stress‐induced apoptosis and microbial infection. Front Immunol. 2019;9:3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jia H, Liu N, Zhang Y, Wang C, Yang Y, Wu Z. 3‐acetyldeoxynivalenol induces cell death through endoplasmic reticulum stress in mouse liver. Environ Pollut. 2021;286:117238. [DOI] [PubMed] [Google Scholar]
- 45. You L, Wang X, Wu W, Nepovimova E, Wu Q, Kuca K. HIF‐1α inhibits T‐2 toxin‐mediated “immune evasion” process by negatively regulating PD‐1/PD‐L1. Toxicology. 2022;480:153324. [DOI] [PubMed] [Google Scholar]
- 46. Qu L, Wang L, Ji H, Fang Y, Lei P, Zhang X, et al. Toxic mechanism and biological detoxification of fumonisins. Toxins. 2022;14:182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kim SH, Singh MP, Sharma C, Kang SC. Fumonisin B1 actuates oxidative stress‐associated colonic damage via apoptosis and autophagy activation in murine model. J Biochem Mol Toxicol. 2018;32:e22161. [DOI] [PubMed] [Google Scholar]
- 48. Singh MP, Kang SC. Endoplasmic reticulum stress‐mediated autophagy activation attenuates fumonisin B1‐induced hepatotoxicity in vitro and in vivo. Food Chem Toxicol. 2017;110:371–382. [DOI] [PubMed] [Google Scholar]
- 49. Song Y, Liu W, Zhao Y, Zang J, Gao H. Fumonisin B1 exposure induces apoptosis of human kidney tubular epithelial cells through regulating PTEN/PI3K/AKT signaling pathway via disrupting lipid raft formation. Toxicon. 2021;204:31–36. [DOI] [PubMed] [Google Scholar]
- 50. Arumugam T, Ghazi T, Chuturgoon A. Fumonisin B1 epigenetically regulates PTEN expression and modulates DNA damage checkpoint regulation in HepG2 liver cells. Toxins. 2020;12:625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gopee NV, He Q, Sharma RP. Fumonisin B1‐induced apoptosis is associated with delayed inhibition of protein kinase C, nuclear factor‐κB and tumor necrosis factor α in LLC‐PK1 cells. Chem Biol Interact. 2003;146:131–145. [DOI] [PubMed] [Google Scholar]
- 52. Chen J, Yang S, Huang S, Yan R, Wang M, Chen S, et al. Transcriptome study reveals apoptosis of porcine kidney cells induced by fumonisin B1 via TNF signalling pathway. Food Chem Toxicol. 2020;139:111274. [DOI] [PubMed] [Google Scholar]
- 53. Han X, Huangfu B, Xu T, Xu W, Asakiya C, Huang K, et al. Research progress of safety of zearalenone: a review. Toxins. 2022;14:386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jing S, Liu C, Zheng J, Dong Z, Guo N. Toxicity of zearalenone and its nutritional intervention by natural products. Food Funct. 2022;13:10374–10400. [DOI] [PubMed] [Google Scholar]
- 55. Parveen M, Zhu Y, Kiyama R. Expression profiling of the genes responding to zearalenone and its analogues using estrogen‐responsive genes. FEBS Lett. 2009;583:2377–2384. [DOI] [PubMed] [Google Scholar]
- 56. Zheng W, Wang B, Li X, Wang T, Zou H, Gu J, et al. Zearalenone promotes cell proliferation or causes cell death? Toxins. 2018;10:184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Abid‐Essefi S, Baudrimont I, Hassen W, Ouanes Z, Mobio TA, Anane R, et al. DNA fragmentation, apoptosis and cell cycle arrest induced by zearalenone in cultured DOK, Vero and Caco‐2 cells: prevention by vitamin E. Toxicology. 2003;192:237–248. [DOI] [PubMed] [Google Scholar]
- 58. Ouanes‐Ben Othmen Z, Essefi S, Bacha H. Mutagenic and epigenetic mechanisms of zearalenone: prevention by vitamin E. World Mycotoxin J. 2008;1:369–374. [Google Scholar]
- 59. Zheng W, Huang Q, Pan S, Fan W, Wang G, Yuan Y, et al. Regulation of oncogenes and gap junction intercellular communication during the proliferative response of zearalenone in TM3 cells. Hum Exp Toxicol. 2016;36:701–708. [DOI] [PubMed] [Google Scholar]
- 60. Wang B, Zheng W, Feng N, Wang T, Zou H, Gu J, et al. The effects of autophagy and PI3K/AKT/m‐TOR signaling pathway on the cell‐cycle arrest of rats primary sertoli cells induced by zearalenone. Toxins. 2018;10:398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Li N, Liu XL, Zhang FL, Tian Y, Zhu M, Meng LY, et al. Whole‐transcriptome analysis of the toxic effects of zearalenone exposure on ceRNA networks in porcine granulosa cells. Environ Pollut. 2020;261:114007. [DOI] [PubMed] [Google Scholar]
- 62. Liu XL, Wu RY, Sun XF, Cheng SF, Zhang RQ, Zhang TY, et al. Mycotoxin zearalenone exposure impairs genomic stability of swine follicular granulosa cells in vitro. Int J Biol Sci. 2018;14:294–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yan R, Wang H, Zhu J, Wang T, Nepovimova E, Long M, et al. Procyanidins inhibit zearalenone‐induced apoptosis and oxidative stress of porcine testis cells through activation of Nrf2 signaling pathway. Food Chem Toxicol. 2022;165:113061. [DOI] [PubMed] [Google Scholar]
- 64. Lin X, Zhu L, Gao X, Kong L, Huang Y, Zhao H, et al. Ameliorative effect of betulinic acid against zearalenone exposure triggers testicular dysfunction and oxidative stress in mice via p38/ERK MAPK inhibition and Nrf2‐mediated antioxidant defense activation. Ecotoxicol Environ Saf. 2022;238:113561. [DOI] [PubMed] [Google Scholar]
- 65. Zhao J, Hai S, Chen J, Ma L, Rahman SU, Zhao C, et al. Zearalenone induces apoptosis in porcine endometrial stromal cells through JNK signaling pathway based on endoplasmic reticulum stress. Toxins. 2022;14:758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Bai J, Li J, Liu N, Jia H, Si X, Zhou Y, et al. Zearalenone induces apoptosis and autophagy by regulating endoplasmic reticulum stress signalling in porcine trophectoderm cells. Anim Nutr. 2023;12:186–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zhang Y, Hu B, Wang M, Tong J, Pan J, Wang N, et al. Selenium protects against zearalenone‐induced oxidative stress and apoptosis in the mouse kidney by inhibiting endoplasmic reticulum stress. Oxid Med Cell Longevity. 2020;2020:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lee H‐J, Oh S‐Y, Jo I. Zearalenone induces endothelial cell apoptosis through activation of a cytosolic Ca2+/ERK1/2/p53/Caspase 3 signaling pathway. Toxins. 2021;13:187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yoon JE, Lee KY, Seok JS, Cheng WN, Kwon HC, Jeong CH, et al. Zearalenone induces endoplasmic reticulum stress and modulates the expression of phase I/II enzymes in human liver cells. Toxins. 2019;12:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Cai G, Si M, Li X, Zou H, Gu J, Yuan Y, et al. Zearalenone induces apoptosis of rat sertoli cells through Fas‐Fas ligand and mitochondrial pathway. Environ Toxicol. 2019;34:424–433. [DOI] [PubMed] [Google Scholar]
- 71. Tang M, Yuan D, Liao P. Berberine improves intestinal barrier function and reduces inflammation, immunosuppression, and oxidative stress by regulating the NF‐κB/MAPK signaling pathway in deoxynivalenol‐challenged piglets. Environ Pollut. 2021;289:117865. [DOI] [PubMed] [Google Scholar]
- 72. Li J, Bai Y, Ma K, Ren Z, Li J, Zhang J, et al. Dihydroartemisinin alleviates deoxynivalenol‐induced liver apoptosis and inflammation in piglets. Ecotoxicol Environ Saf. 2022;241:113811. [DOI] [PubMed] [Google Scholar]
- 73. Chen J, Wang M, Wang H, Long M. Zearalenone promotes apoptosis of mouse Leydig cells by targeting phosphatase and tensin homolog and thus inhibiting the PI3K/AKT signal pathway. Environ Sci Pollut Res. 2021;28:67779–67787. [DOI] [PubMed] [Google Scholar]
- 74. EFSA Panel on Contaminants in the Food Chain (CONTAM), Knutsen HK, Alexander J, Barregard L, Bignami M, Bruschweiler B, et al. Risks for animal health related to the presence of zearalenone and its modified forms in feed. EFSA J. 2017;15:4851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. IPCC . Climate change 2023: synthesis report. Geneva(CH): IPCC; 2023. [Google Scholar]
- 76. Medina A, Akbar A, Baazeem A, Rodriguez A, Magan N. Climate change, food security and mycotoxins: do we know enough? Fungal Biol Rev. 2017;31:143–154. [Google Scholar]
- 77. Paterson RRM, Lima N. How will climate change affect mycotoxins in food? Food Res Int. 2010;43:1902–1914. [Google Scholar]
- 78. Isebaert S, De Saeger S, Devreese R, Verhoeven R, Maene P, Heremans B, et al. Mycotoxin‐producing Fusarium species occurring in winter wheat in Belgium (Flanders) during 2002‐2005. J Phytopath. 2009;157:108–116. [Google Scholar]
- 79. Gruber‐Dorninger C, Jenkins T, Schatzmayr G. Global mycotoxin occurrence in feed: a ten‐year survey. Toxins. 2019;11:375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Sanchis V, Magan N. Environmental conditions affecting mycotoxins. In: Magan N, Olsen M editors. Mycotoxins in food: detection and control. Abington: CRC Press LLC; 2004. p. 174–189 [Google Scholar]
- 81. Yu S, Jia B, Li K, Zhou H, Lai W, Tang Y, et al. Pre‐warning of abiotic factors in maize required for potential contamination of Fusarium mycotoxins via response surface analysis. Food Control. 2021;121:107570. [Google Scholar]
- 82. Paterson RRM, Lima N. Further mycotoxin effects from climate change. Food Res Int. 2011;44:2555–2566. [Google Scholar]
- 83. Santiago R, Cao A, Butrón A. Genetic factors involved in fumonisin accumulation in maize kernels and their implications in maize agronomic management and breeding. Toxins. 2015;7:3267–3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Wu F, Bhatnagar D, Bui‐Klimke T, Carbone I, Hellmich R, Munkvold G, et al. Climate change impacts on mycotoxin risks in US maize. World Mycotoxin J. 2011;4:79–93. [Google Scholar]
- 85. Shelby RA. Differential fumonisin production in maize hybrids. Plant Dis. 1994;78:582–584. [Google Scholar]
- 86. Torres OA, Palencia E, de Pratdesaba LL, Grajeda R, Fuentes M, Speer MC, et al. Estimated fumonisin exposure in Guatemala is greatest in consumers of lowland maize. J Nutr. 2007;137:2723–2729. [DOI] [PubMed] [Google Scholar]
- 87. Atukwase A, Kaaya AN, Muyanja C. Factors associated with fumonisin contamination of maize in Uganda. J Sci Food Agric. 2009;89:2393–2398. [Google Scholar]
- 88. van der Fels‐Klerx HJ, Olesen JE, Madsen MS, Goedhart PW. Climate change increases deoxynivalenol contamination of wheat in north‐western Europe. Food Addit Contam Part A. 2012;29:1593–1604. [DOI] [PubMed] [Google Scholar]
- 89. Miller JD, Miller JD. Factors that affect the occurrence of fumonisin. Environ Health Perspect. 2001;109:321–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Akello J, Ortega‐Beltran A, Katati B, Atehnkeng J, Augusto J, Mwila CM, et al. Prevalence of aflatoxin‐ and fumonisin‐producing fungi associated with cereal crops grown in Zimbabwe and their associated risks in a climate change scenario. Foods. 2021;10:287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Salvacion AR, Pangga IB, Cumagun CJR. Assessment of mycotoxin risk on corn in the Philippines under current and future climate change conditions. Rev Environ Health. 2015;30:135–142. [DOI] [PubMed] [Google Scholar]
- 92. Janić Hajnal E, Kos J, Radić B, Anić M, Radović R, Kudumija N, et al. Impact of climate changes on the natural prevalence of Fusarium mycotoxins in maize harvested in Serbia and Croatia. Foods. 2023;12:1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Campa R, Hooker DC, Miller JD, Schaafsma AW, Hammond BG. Modeling effects of environment, insect damage, and Bt genotypes on fumonisin accumulation in maize in Argentina and the Philippines. Mycopathologia. 2005;159:539–552. [DOI] [PubMed] [Google Scholar]
- 94. Hooker DC, Schaafsma AW, Tamburic‐Ilincic L. Using weather variables pre‐and post‐heading to predict deoxynivalenol content in winter wheat. Plant Dis. 2002;86:611–619. [DOI] [PubMed] [Google Scholar]
- 95. Birr T, Verreet J‐A, Klink H. Prediction of deoxynivalenol and zearalenone in winter wheat grain in a maize‐free crop rotation based on cultivar susceptibility and meteorological factors. J Plant Dis Protect. 2018;126:13–27. [Google Scholar]
- 96. Joo Y, Ok HE, Kim J, Lee SY, Jang SK, Park KH, et al. A statistical model for determining zearalenone contamination in rice (Oryza sativa L.) at harvest and its prediction under different climate change scenarios in South Korea. Appl Biol Chem. 2019;62:38. [Google Scholar]
- 97. Toreti A, Deryng D, Tubiello FN, Müller C, Kimball BA, Moser G, et al. Narrowing uncertainties in the effects of elevated CO2 on crops. Nat Food. 2020;1:775–782. [DOI] [PubMed] [Google Scholar]
- 98. Vaughan MM, Huffaker A, Schmelz EA, Dafoe NJ, Christensen S, Sims J, et al. Effects of elevated [CO2] on maize defence against mycotoxigenic Fusarium verticillioides . Plant Cell Environ. 2014;37:2691–2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Vaughan MM, Huffaker A, Schmelz EA, Dafoe NJ, Christensen SA, McAuslane HJ, et al. Interactive effects of elevated [CO2] and drought on the maize phytochemical defense response against mycotoxigenic Fusarium verticillioides . PLoS One. 2016;11:e0159270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Hay WT, McCormick SP, Vaughan MM. Effects of atmospheric CO2 and temperature on wheat and corn susceptibility to Fusarium graminearum and deoxynivalenol contamination. Plants. 2021;10:2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Bencze S, Puskás K, Vida G, Karsai I, Balla K, Komáromi J, et al. Rising atmospheric CO2 concentration may imply higher risk of Fusarium mycotoxin contamination of wheat grains. Mycotoxin Res. 2017;33:229–236. [DOI] [PubMed] [Google Scholar]
- 102. Kahla A, Verheecke‐Vaessen C, Delpino‐Deelias M, Gutierrez‐Pozo M, Medina A, Magan N, et al. Acclimatisation of Fusarium langsethiae, F. poae and F. sporotrichioides to elevated CO2: impact on fungal growth and mycotoxin production on oat‐based media. Int J Food Microbiol. 2023;394:110176. [DOI] [PubMed] [Google Scholar]
- 103. Singh J, Mehta A. Rapid and sensitive detection of mycotoxins by advanced and emerging analytical methods: a review. Food Sci Nutr. 2020;8:2183–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Dall'Asta C, Mangia M, Berthiller F, Molinelli A, Sulyok M, Schuhmacher R, et al. Difficulties in fumonisin determination: the issue of hidden fumonisins. Anal Bioanal Chem. 2009;395:1335–1345. [DOI] [PubMed] [Google Scholar]
- 105. Spanjer MC, Rensen PM, Scholten JM. LC–MS/MS multi‐method for mycotoxins after single extraction, with validation data for peanut, pistachio, wheat, maize, cornflakes, raisins and figs. Food Addit Contam Part A. 2008;25:472–489. [DOI] [PubMed] [Google Scholar]
- 106. Tevell Åberg A, Solyakov A, Bondesson U. Development and in‐house validation of an LC‐MS/MS method for the quantification of the mycotoxins deoxynivalenol, zearalenone, T‐2 and HT‐2 toxin, ochratoxin A and fumonisin B1 and B2 in vegetable animal feed. Food Addit Contam Part A. 2013;30:541–549. [DOI] [PubMed] [Google Scholar]
- 107. Varga E, Fodor P, Soros C. Multi‐mycotoxin LC‐MS/MS method validation and its application to fifty‐four wheat flours in Hungary. Food Addit Contam Part A. 2021;38:670–680. [DOI] [PubMed] [Google Scholar]
- 108. Cendoya E, Nichea MJ, Monge MP, Sulyok M, Chiacchiera SM, Ramirez ML. Fumonisin occurrence in wheat‐based products from Argentina. Food Addit Contam Part B. 2018;12:31–37. [DOI] [PubMed] [Google Scholar]
- 109. Wang Y, Xiao C, Guo J, Yuan Y, Wang J, Liu L, et al. Development and application of a method for the analysis of 9 mycotoxins in maize by HPLC‐MS/MS. J Food Sci. 2013;78:M1752–M1757. [DOI] [PubMed] [Google Scholar]
- 110. Ueno Y, Iijima K, Wang S‐D, Sugiura Y, Sekijima M, Tanaka T, et al. Fumonisins as a possible contributory risk factor for primary liver cancer: a 3‐year study of corn harvested in Haimen, China, by HPLC and ELISA. Food Chem Toxicol. 1997;35:1143–1150. [DOI] [PubMed] [Google Scholar]
- 111. De La Campa R, Miller JD, Hendricks K. Fumonisin in tortillas produced in small‐scale facilities and effect of traditional masa production methods on this mycotoxin. J Agricult Food Chem. 2004;52:4432–4437. [DOI] [PubMed] [Google Scholar]
- 112. Kaltner F, Rampl C, Rychlik M, Zimmermann T, Rohe A. Development and validation of a cost‐effective HPLC‐FLD method for routine analysis of fumonisins B1 and B2 in corn and corn products. Food Anal Methods. 2016;10:1349–1358. [Google Scholar]
- 113. Kirimker SE, Turksoy S, Kabak B. Assessment of dietary exposure to deoxynivalenol and fumonisin in the population of infants and toddlers in Turkey. Food Chem Toxicol. 2020;140:111304. [DOI] [PubMed] [Google Scholar]
- 114. Zhao XS, Kong WJ, Wang S, Wei JH, Yang MH. Simultaneous analysis of multiple mycotoxins in Alpinia oxyphylla by UPLC‐MS/MS. World Mycotoxin J. 2017;10:41–51. [Google Scholar]
- 115. Leite M, Freitas A, Barbosa J, Ramos F. Comprehensive assessment of different extraction methodologies for optimization and validation of an analytical multi‐method for determination of emerging and regulated mycotoxins in maize by UHPLC‐MS/MS. Food Chem Adv. 2023;2:100145. [Google Scholar]
- 116. Ling S, Pang J, Yu J, Wang R, Liu L, Ma Y, et al. Preparation and identification of monoclonal antibody against fumonisin B1 and development of detection by Ic‐ELISA. Toxicon. 2014;80:64–72. [DOI] [PubMed] [Google Scholar]
- 117. Shu M, Xu Y, Dong J, Zhong C, Hammock BD, Wang W, et al. Development of a noncompetitive idiometric nanobodies phage immumoassay for the determination of fumonisin B1. Food Agric Immunol. 2019;30:510–521. [Google Scholar]
- 118. Wang XC, Bao M, Li FH, Fan HX, Li HS, Li Y, et al. Development of a sensitive, competitive, indirect ELISA for the detection of fumonisin B1 in corn originating from Anhui province, China. J Environ Sci Health Part B. 2015;51:107–112. [DOI] [PubMed] [Google Scholar]
- 119. Liu X, Xu Y, He Q, He Z, Xiong Z. Application of mimotope peptides of fumonisin b1 in peptide ELISA. J Agricult Food Chem. 2013;61:4765–4770. [DOI] [PubMed] [Google Scholar]
- 120. Ren W, Xu Y, Huang Z, Li Y, Tu Z, Zou L, et al. Single‐chain variable fragment antibody‐based immunochromatographic strip for rapid detection of fumonisin B1 in maize samples. Food Chem. 2020;319:126546. [DOI] [PubMed] [Google Scholar]
- 121. Zha C, An X, Zhang J, Wei L, Zhang Q, Yang Q, et al. Indirect signal amplification strategy with a universal probe‐based lateral flow immunoassay for the rapid quantitative detection of fumonisin B1. Anal Methods. 2022;14:708–716. [DOI] [PubMed] [Google Scholar]
- 122. Huang X, Huang X, Xie J, Li X, Huang Z. Rapid simultaneous detection of fumonisin B1 and deoxynivalenol in grain by immunochromatographic test strip. Anal Biochem. 2020;606:113878. [DOI] [PubMed] [Google Scholar]
- 123. Huang X, Huang T, Li X, Huang Z. Flower‐like gold nanoparticles‐based immunochromatographic test strip for rapid simultaneous detection of fumonisin B1 and deoxynivalenol in Chinese traditional medicine. J Pharm Biomed Anal. 2020;177:112895. [DOI] [PubMed] [Google Scholar]
- 124. Munawar H, Smolinska‐Kempisty K, Cruz AG, Canfarotta F, Piletska E, Karim K, et al. Molecularly imprinted polymer nanoparticle‐based assay (MINA): application for fumonisin B1 determination. Analyst (Lond). 2018;143:3481–3488. [DOI] [PubMed] [Google Scholar]
- 125. Munawar H, Garcia‐Cruz A, Majewska M, Karim K, Kutner W, Piletsky SA. Electrochemical determination of fumonisin B1 using a chemosensor with a recognition unit comprising molecularly imprinted polymer nanoparticles. Sens Actuators B. 2020;321:128552. [Google Scholar]
- 126. Zhang W, Xiong H, Chen M, Zhang X, Wang S. Surface‐enhanced molecularly imprinted electrochemiluminescence sensor based on Ru@SiO2 for ultrasensitive detection of fumonisin B1 . Biosens Bioelectron. 2017;96:55–61. [DOI] [PubMed] [Google Scholar]
- 127. Mao L, Ji K, Yao L, Xue X, Wen W, Zhang X, et al. Molecularly imprinted photoelectrochemical sensor for fumonisin B1 based on GO‐CdS heterojunction. Biosens Bioelectron. 2019;127:57–63. [DOI] [PubMed] [Google Scholar]
- 128. Lu L, Seenivasan R, Wang Y‐C, Yu J‐H, Gunasekaran S. An electrochemical immunosensor for rapid and sensitive detection of mycotoxins fumonisin b1 and deoxynivalenol. Electrochim Acta. 2016;213:89–97. [Google Scholar]
- 129. Yang X, Zhou X, Zhang X, Qing Y, Luo M, Liu X, et al. A highly sensitive electrochemical immunosensor for fumonisin B1 detection in corn using single‐walled carbon nanotubes/chitosan. Electroanalysis. 2015;27:2679–2687. [Google Scholar]
- 130. Wei M, Xin L, Feng S, Liu Y. Simultaneous electrochemical determination of ochratoxin A and fumonisin B1 with an aptasensor based on the use of a Y‐shaped DNA structure on gold nanorods. Microchim Acta. 2020;187:102. [DOI] [PubMed] [Google Scholar]
- 131. Dong N, Li Y, Meng S, Liu S, Liu Y, Liu D, et al. Tetrahedral DNA nanostructure‐based ratiometric electrochemical aptasensor for fumonisin B1: a unity of opposites in binding site and steric hindrance of large‐sized DNA for signal amplification. Sens Actuators B. 2023;394:134341. [Google Scholar]
- 132. Sun Y, Lv Y, Zhang Y, Wang Z. A stimuli‐responsive colorimetric aptasensor based on the DNA hydrogel‐coated MOF for fumonisin B1 determination in food samples. Food Chem. 2023;403:134242. [DOI] [PubMed] [Google Scholar]
- 133. Zhao Y, Luo Y, Li T, Song Q. Au NPs driven electrochemiluminescence aptasensors for sensitive detection of fumonisin B1. RSC Adv. 2014;4:57709–57714. [Google Scholar]
- 134. Bertuzzi T, Camardo Leggieri M, Battilani P, Pietri A. Co‐occurrence of type A and B trichothecenes and zearalenone in wheat grown in northern Italy over the years 2009–2011. Food Addit Contamin Part B. 2014;7:273–281. [DOI] [PubMed] [Google Scholar]
- 135. Rasmussen RR, Storm IMLD, Rasmussen PH, Smedsgaard J, Nielsen KF. Multi‐mycotoxin analysis of maize silage by LC‐MS/MS. Anal Bioanal Chem. 2010;397:765–776. [DOI] [PubMed] [Google Scholar]
- 136. Tahoun IF, Gab‐Allah MA, Yamani RN, Shehata AB. Development and validation of a reliable LC‐MS/MS method for simultaneous determination of deoxynivalenol and T‐2 toxin in maize and oats. Microchem J. 2021;169:106599. [Google Scholar]
- 137. Golge O, Kabak B. Occurrence of deoxynivalenol and zearalenone in cereals and cereal products from Turkey. Food Control. 2020;110:106982. [Google Scholar]
- 138. Han L, Li Y‐T, Jiang J‐Q, Li R‐F, Fan G‐Y, Lv J‐M, et al. Development of a direct competitive ELISA kit for detecting deoxynivalenol contamination in wheat. Molecules. 2019;25:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Li M, Sun M, Hong X, Duan J, Du D. Survey of deoxynivalenol contamination in agricultural products in the Chinese market using an ELISA kit. Toxins. 2018;11:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Li Y, Liu G, Fu X, He J, Wang Z, Hou J, et al. High‐sensitive chemiluminescent ELISA method investigation for the determination of deoxynivalenol in rice. Food Anal Methods. 2014;8:656–660. [Google Scholar]
- 141. Yan T, Zhang Q, Wang D, Li P, Tang X, Zhang W. Determination of deoxynivalenol by ELISA and mmunochromatographic strip assay based on monoclonal antibodies. Toxin Rev. 2019;40:285–291. [Google Scholar]
- 142. Huang Z‐B, Xu Y, Li L‐S, Li Y‐P, Zhang H, He Q‐H. Development of an immunochromatographic strip test for the rapid simultaneous detection of deoxynivalenol and zearalenone in wheat and maize. Food Control. 2012;28:7–12. [Google Scholar]
- 143. Xu Y, Huang Z‐B, He Q‐H, Deng S‐Z, Li L‐S, Li Y‐P. Development of an immunochromatographic strip test for the rapid detection of deoxynivalenol in wheat and maize. Food Chem. 2010;119:834–839. [Google Scholar]
- 144. Wei T, Ren P, Huang L, Ouyang Z, Wang Z, Kong X, et al. Simultaneous detection of aflatoxin B1, ochratoxin A, zearalenone and deoxynivalenol in corn and wheat using surface plasmon resonance. Food Chem. 2019;300:125176. [DOI] [PubMed] [Google Scholar]
- 145. Valera E, García‐Febrero R, Elliott CT, Sánchez‐Baeza F, Marco MP. Electrochemical nanoprobe‐based immunosensor for deoxynivalenol mycotoxin residues analysis in wheat samples. Anal Bioanal Chem. 2019;411:1915–1926. [DOI] [PubMed] [Google Scholar]
- 146. Radi A‐E, Eissa A, Wahdan T. Impedimetric sensor for deoxynivalenol based on electropolymerised molecularly imprinted polymer on the surface of screen‐printed gold electrode. Int J Environ Anal Chem. 2019;101:2586–2597. [Google Scholar]
- 147. Li W, Diao K, Qiu D, Zeng Y, Tang K, Zhu Y, et al. A highly‐sensitive and selective antibody‐like sensor based on molecularly imprinted poly(L‐arginine) on COOH‐MWCNTs for electrochemical recognition and detection of deoxynivalenol. Food Chem. 2021;350:129229. [DOI] [PubMed] [Google Scholar]
- 148. Choi S‐W, Chang H‐J, Lee N, Chun HS. A surface plasmon resonance sensor for the detection of deoxynivalenol using a molecularly imprinted polymer. Sensors. 2011;11:8654–8664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Zhao X, Shen H, Huo B, Wang Y, Gao Z. A novel bionic magnetic SERS aptasensor for the ultrasensitive detection of deoxynivalenol based on “dual antennae” nano‐silver. Biosens Bioelectron. 2022;211:114383. [DOI] [PubMed] [Google Scholar]
- 150. Yu W, Lin X, Duan N, Wang Z, Wu S. A fluorescence and surface‐enhanced Raman scattering dual‐mode aptasensor for sensitive detection of deoxynivalenol based on gold nanoclusters and silver nanoparticles modified metal‐polydopamine framework. Anal Chim Acta. 2023;1244:340846. [DOI] [PubMed] [Google Scholar]
- 151. Duan N, Li C, Song M, Ren K, Wang Z, Wu S. Deoxynivalenol fluorescence aptasensor based on AuCu bimetallic nanoclusters and MoS2. Microchim Acta. 2022;189:296. [DOI] [PubMed] [Google Scholar]
- 152. Wang K, He B, Xie L, Li L, Yang J, Liu R, et al. Exonuclease III‐assisted triple‐amplified electrochemical aptasensor based on PtPd NPs/PEI‐rGO for deoxynivalenol detection. Sens Actuators, B. 2021;349:130767. [Google Scholar]
- 153. Urraca JL, Marazuela MD, Moreno‐Bondi MC. Analysis for zearalenone and α‐zearalenol in cereals and swine feed using accelerated solvent extraction and liquid chromatography with fluorescence detection. Anal Chim Acta. 2004;524:175–183. [Google Scholar]
- 154. Pascari X, Weigel S, Marin S, Sanchis V, Maul R. Detection and quantification of zearalenone and its modified forms in enzymatically treated oat and wheat flour. J Food Sci Technol. 2023;60:1367–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Pochivalov A, Pavlova K, Garmonov S, Bulatov A. Behaviour of deep eutectic solvent based on terpenoid and long‐chain alcohol during dispersive liquid‐liquid microextraction: determination of zearalenone in cereal samples. J Mol Liq. 2022;366:120231. [Google Scholar]
- 156. Rahmani A, Jinap S, Soleimany F. Validation of the procedure for the simultaneous determination of aflatoxins ochratoxin A and zearalenone in cereals using HPLC‐FLD. Food Addit Contam Part A. 2010;27:1683–1693. [DOI] [PubMed] [Google Scholar]
- 157. Pei S‐C, Lee W‐J, Zhang G‐P, Hu X‐F, Eremin SA, Zhang L‐J. Development of anti‐zearalenone monoclonal antibody and detection of zearalenone in corn products from China by ELISA. Food Control. 2013;31:65–70. [Google Scholar]
- 158. Zhao F, Shen Q, Wang H, Han X, Yang Z. Development of a rapid magnetic bead‐based immunoassay for sensitive detection of zearalenone. Food Control. 2017;79:227–233. [Google Scholar]
- 159. Thongrussamee T, Kuzmina NS, Shim WB, Jiratpong T, Eremin SA, Intrasook J, et al. Monoclonal‐based enzyme‐linked immunosorbent assay for the detection of zearalenone in cereals. Food Addit Contamin Part A. 2008;25:997–1006. [DOI] [PubMed] [Google Scholar]
- 160. Jiang X, Li X, Yang Z, Eremin SA, Zhang X. Evaluation and optimization of three different immunoassays for rapid detection zearalenone in fodders. Food Anal Methods. 2016;10:256–262. [Google Scholar]
- 161. Duan H, Chen X, Xu W, Fu J, Xiong Y, Wang A. Quantum‐DoT submicrobead‐based immunochromatographic assay for quantitative and sensitive detection of zearalenone. Talanta. 2015;132:126–131. [DOI] [PubMed] [Google Scholar]
- 162. Székács I, Adányi N, Szendrő I, Székács A. Direct and competitive optical grating immunosensors for determination of Fusarium mycotoxin zearalenone. Toxins. 2021;13:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Foubert A, Beloglazova NV, Hedström M, De Saeger S. Antibody immobilization strategy for the development of a capacitive immunosensor detecting zearalenone. Talanta. 2019;191:202–208. [DOI] [PubMed] [Google Scholar]
- 164. Choi S‐W, Chang H‐J, Lee N, Kim J‐H, Chun HS. Detection of mycoestrogen zearalenone by a molecularly imprinted polypyrrole‐based surface plasmon resonance (SPR) sensor. J Agricult Food Chem. 2009;57:1113–1118. [DOI] [PubMed] [Google Scholar]
- 165. Yarynka D, Chegel V, Piletska E, Piletsky S, Dubey L, Dubey I, et al. An enhanced fluorescent sensor system based on molecularly imprinted polymer chips with silver nanoparticles for highly‐sensitive zearalenone analysis. Analyst. 2023;148:2633–2643. [DOI] [PubMed] [Google Scholar]
- 166. Radi AE, Eissa A, Wahdan T. Molecularly imprinted impedimetric sensor for determination of mycotoxin zearalenone. Electroanalysis. 2020;32:1788–1794. [Google Scholar]
- 167. Ma L, Bai L, Zhao M, Zhou J, Chen Y, Mu Z. An electrochemical aptasensor for highly sensitive detection of zearalenone based on PEI‐MoS2‐MWCNTs nanocomposite for signal enhancement. Anal Chim Acta. 2019;1060:71–78. [DOI] [PubMed] [Google Scholar]
- 168. Guo Z, Gao L, Yin L, Arslan M, El‐Seedi HR, Zou X. Novel mesoporous silica surface loaded gold nanocomposites SERS aptasensor for sensitive detection of zearalenone. Food Chem. 2023;403:134384. [DOI] [PubMed] [Google Scholar]
- 169. Wu Z, Xu E, Chughtai MFJ, Jin Z, Irudayaraj J. Highly sensitive fluorescence sensing of zearalenone using a novel aptasensor based on upconverting nanoparticles. Food Chem. 2017;230:673–680. [DOI] [PubMed] [Google Scholar]
- 170. Capriotti AL, Caruso G, Cavaliere C, Foglia P, Samperi R, Laganà A. Multiclass mycotoxin analysis in food, environmental and biological matrices with chromatography/mass spectrometry. Mass Spectrom Rev. 2012;31:466–503. [DOI] [PubMed] [Google Scholar]
- 171. Mbundi L, Gallar‐Ayala H, Khan MR, Barber JL, Losada S, Busquets R. Advances in the analysis of challenging food contaminants. In: Fishbein JC, Heilman JM editors. Advances in molecular toxicology. London, UK: Elsevier; 2014. p. 35–105. [Google Scholar]
- 172. Steiner D, Sulyok M, Malachová A, Mueller A, Krska R. Realizing the simultaneous liquid chromatography‐tandem mass spectrometry‐based quantification of >1200 biotoxins, pesticides and veterinary drugs in complex feed. J Chromatogr A. 2020;1629:461502. [DOI] [PubMed] [Google Scholar]
- 173. Tittlemier SA, Brunkhorst J, Cramer B, DeRosa MC, Lattanzio VMT, Malone R, et al. Developments in mycotoxin analysis: an update for 2019‐2020. World Mycotoxin J. 2021;14:3–26. [Google Scholar]
- 174. Tittlemier SA, Cramer B, Dall'Asta C, DeRosa MC, Lattanzio VMT, Malone R, et al. Developments in mycotoxin analysis: an update for 2020‐2021. World Mycotoxin J. 2022;15:3–25. [Google Scholar]
- 175. Mateus ARS, Barros S, Pena A, Silva AS. Development and validation of QuEChERS followed by UHPLC‐ToF‐MS method for determination of multi‐mycotoxins in pistachio nuts. Molecules. 2021;26:5754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Pi J, Jin P, Zhou S, Wang L, Wang H, Huang J, et al. Combination of ultrasonic‐assisted aqueous two‐phase extraction with solidifying organic drop‐dispersive liquid–liquid microextraction for simultaneous determination of nine mycotoxins in medicinal and edible foods by HPLC with in‐series DAD and FLD. Food Anal Methods. 2022;15:428–439. [Google Scholar]
- 177. Lee SY, Woo SY, Malachová A, Michlmayr H, Kim SH, Kang GJ, et al. Simple validated method for simultaneous determination of deoxynivalenol, nivalenol, and their 3‐β‐D‐glucosides in baby formula and Korean rice wine via HPLC‐UV with immunoaffinity cleanup. Food Addit Contam Part A. 2019;36:964–975. [DOI] [PubMed] [Google Scholar]
- 178. Koczula KM, Gallotta A. Lateral flow assays. Essays Biochem. 2016;60:111–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Wang Y, Zhang C, Wang J, Knopp D. Recent progress in rapid determination of mycotoxins based on emerging biorecognition molecules: a review. Toxins. 2022;14:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Xing K‐Y, Shan S, Liu D‐F, Lai W‐H. Recent advances of lateral flow immunoassay for mycotoxins detection. Trends Anal Chem. 2020;133:116087. [Google Scholar]
- 181. Liu Z, Hua Q, Wang J, Liang Z, Li J, Wu J, et al. A smartphone‐based dual detection mode device integrated with two lateral flow immunoassays for multiplex mycotoxins in cereals. Biosens Bioelectron. 2020;158:112178. [DOI] [PubMed] [Google Scholar]
- 182. Li R, Wen Y, Wang F, He P. Recent advances in immunoassays and biosensors for mycotoxins detection in feedstuffs and foods. J Anim Sci Biotechnol. 2021;12:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Jia M, Liao X, Fang L, Jia B, Liu M, Li D, et al. Recent advances on immunosensors for mycotoxins in foods and other commodities. Trends Anal Chem. 2021;136:116193. [Google Scholar]
- 184. Aydin M, Aydin EB, Sezginturk MK. Advances in immunosensor technology. In: Makowski G, editor. Advances in clinical chemistry. London, UK: Elsevier; 2021. p. 3–50. [DOI] [PubMed] [Google Scholar]
- 185. Yang M, Zhang Y, Cui M, Tian Y, Zhang S, Peng K, et al. A smartphone‐based quantitative detection platform of mycotoxins based on multiple‐color upconversion nanoparticles. Nanoscale. 2018;10:15865–15874. [DOI] [PubMed] [Google Scholar]
- 186. Li Y, Chen Q, Xu X, Jin Y, Wang Y, Zhang L, et al. Microarray surface enhanced Raman scattering based immunosensor for multiplexing detection of mycotoxin in foodstuff. Sens Actuators, B. 2018;266:115–123. [Google Scholar]
- 187. Evtugyn G, Hianik T. Electrochemical immuno‐ and aptasensors for mycotoxin determination. Chemosensors. 2019;7:10. [Google Scholar]
- 188. Xu Y, Jiang X, Zhou Y, Ma M, Wang M, Ying B. Systematic evolution of ligands by exponential enrichment technologies and aptamer‐based applications: recent progress and challenges in precision medicine of infectious diseases. Front Bioeng Biotechnol. 2021;9:704077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Munzar JD, Ng A, Juncker D. Duplexed aptamers: history, design, theory, and application to biosensing. Chem Soc Rev. 2019;48:1390–1419. [DOI] [PubMed] [Google Scholar]
- 190. Ronkainen NJ, Halsall HB, Heineman WR. Electrochemical biosensors. Chem Soc Rev. 2010;39:1747–1763. [DOI] [PubMed] [Google Scholar]
- 191. Zhang Y, He B, Zhao R, Bai C, Zhang Y, Jin H, et al. Electrochemical aptasensor based on the target‐induced strand displacement strategy‐driven for T‐2 toxin detection. Sci Total Environ. 2022;849:157769. [DOI] [PubMed] [Google Scholar]
- 192. Zhang N, Liu B, Cui X, Li Y, Tang J, Wang H, et al. Recent advances in aptasensors for mycotoxin detection: on the surface and in the colloid. Talanta. 2021;223:121729. [DOI] [PubMed] [Google Scholar]
- 193. Mukunzi D, Habimana JD, Li Z, Zou X. Mycotoxins detection: view in the lens of molecularly imprinted polymer and nanoparticles. Crit Rev Food Sci Nutr. 2022;63:1–35. [DOI] [PubMed] [Google Scholar]
- 194. BelBruno JJ. Molecularly imprinted polymers. Chem Rev. 2019;119:94–119. [DOI] [PubMed] [Google Scholar]
- 195. Sergeyeva T, Yarynka D, Dubey L, Dubey I, Piletska E, Linnik R, et al. Sensor based on molecularly imprinted polymer membranes and smartphone for detection of Fusarium contamination in cereals. Sensors. 2020;20:4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Munawar H, Safaryan AHM, De Girolamo A, Garcia‐Cruz A, Marote P, Karim K, et al. Determination of fumonisin B1 in maize using molecularly imprinted polymer nanoparticles‐based assay. Food Chem. 2019;298:125044. [DOI] [PubMed] [Google Scholar]
- 197. Calahorra‐Rio L, Guadaño‐Sánchez M, Moya‐Cavas T, Urraca JL. Magnetic core‐shell nanoparticles using molecularly imprinted polymers for zearalenone determination. Molecules. 2022;27:8166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Drakopoulos D, Gimeno A, Kägi A, Jenny E, Bänziger I, Musa T, et al. Innovative cropping systems to reduce Fusarium mycotoxins in wheat. Agrarforsch Schweiz. 2021;12:16–23. [Google Scholar]
- 199. Qiu J, Dong F, Yu M, Xu J, Shi J. Effect of preceding crop on Fusarium species and mycotoxin contamination of wheat grains. J Sci Food Agric. 2016;96:4536–4541. [DOI] [PubMed] [Google Scholar]
- 200. Drakopoulos D, Kägi A, Six J, Zorn A, Wettstein FE, Bucheli TD, et al. The agronomic and economic viability of innovative cropping systems to reduce Fusarium head blight and related mycotoxins in wheat. Agricult Sys. 2021;192:103198. [Google Scholar]
- 201. Bernhoft A, Wang J, Leifert C. Effect of organic and conventional cereal production methods on Fusarium head blight and mycotoxin contamination levels. Agronomy. 2022;12:797. [Google Scholar]
- 202. Mesterházy Á, Bartók T, Mirocha CG, Komoróczy R. Nature of wheat resistance to Fusarium head blight and the role of deoxynivalenol for breeding. Plant Breed. 1999;118:97–110. [Google Scholar]
- 203. Jia H, Zhou J, Xue S, Li G, Yan H, Ran C, et al. A journey to understand wheat Fusarium head blight resistance in the Chinese wheat landrace Wangshuibai. Crop J. 2018;6:48–59. [Google Scholar]
- 204. Bai G, Su Z, Cai J. Wheat resistance to Fusarium head blight. Can J Plant Pathol. 2018;40:336–346. [Google Scholar]
- 205. McLaughlin JE, Darwish NI, Garcia‐Sanchez J, Tyagi N, Trick HN, McCormick S, et al. A lipid transfer protein has antifungal and antioxidant activity and suppresses Fusarium head blight disease and DON accumulation in transgenic wheat. Phytopathology.® 2021;111:671–683. [DOI] [PubMed] [Google Scholar]
- 206. Attia S, Khan RS, Iqbal A, Hussain SA, Kamil A, Khan MA, et al. Host‐induced gene silencing: an effective control strategy against Fusarium species. J Plant Dis Protect. 2022;129:1025–1030. [Google Scholar]
- 207. de Chaves MA, Reginatto P, da Costa BS, de Paschoal RI, Teixeira ML, Fuentefria AM. Fungicide resistance in Fusarium graminearum species complex. Curr Microbiol. 2022;79:62. [DOI] [PubMed] [Google Scholar]
- 208. Paul PA, Lipps PE, Hershman DE, McMullen MP, Draper MA, Madden LV. Efficacy of triazole‐based fungicides for Fusarium head blight and deoxynivalenol control in wheat: a multivariate meta‐analysis. Phytopathology®. 2008;98:999–1011. [DOI] [PubMed] [Google Scholar]
- 209. Tang G, Chen Y, Xu JR, Kistler HC, Ma Z. The fungal myosin I is essential for Fusarium toxisome formation. PLoS Pathog. 2018;14:e1006827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Chen H, Wu Q, Zhang G, Wu J, Zhu F, Yang H, et al. Carbendazim‐resistance of Gibberella zeae associated with Fusarium head blight and its management in Jiangsu Province, China. Crop Prot. 2019;124:104866. [Google Scholar]
- 211. Marin S, Sanchis V, Sanz D, Castel I, Ramos AJ, Canela R, et al. Control of growth and fumonisin B1 production by Fusarium verticillioides and Fusarium proliferatum isolates in moist maize with propionate preservatives. Food Addit Contam. 1999;16:555–563. [DOI] [PubMed] [Google Scholar]
- 212. Farnochi MC, Torres AM, Magan N, Chulze SN. Effect of antioxidants and competing mycoflora on Fusarium verticillioides and F. proliferatum populations and fumonisin production on maize grain. J Stored Prod Res. 2005;41:211–219. [Google Scholar]
- 213. Daou R, Joubrane K, Maroun RG, Khabbaz LR, Ismail A, Khoury AE. Mycotoxins: factors influencing production and control strategies. AIMS Agricult Food. 2021;6:416–447. [Google Scholar]
- 214. Chalivendra S, Ham JH. Bacilli in the biocontrol of mycotoxins. In: Islam MT, Rahman MM, Pandey P, Boehme MH, Haesaert G, editors. Bacilli and agrobiotechnology: phytostimulation and biocontrol Cham. Germany: Springer; 2019. p. 49–62. [Google Scholar]
- 215. Salazar B, Ortiz A, Keswani C, Minkina T, Mandzhieva S, Pratap Singh S, et al. Bacillus spp. as bio‐factories for antifungal secondary metabolites: innovation beyond whole organism formulations. Microb Ecol. 2022;86:1–24. [DOI] [PubMed] [Google Scholar]
- 216. Fazle Rabbee M, Baek KH. Antimicrobial activities of lipopeptides and polyketides of Bacillus velezensis for agricultural applications. Molecules. 2020;25:4973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Krishnan N, Velramar B, Velu RK. Investigation of antifungal activity of surfactin against mycotoxigenic phytopathogenic fungus Fusarium moniliforme and its impact in seed germination and mycotoxicosis. Pest Biochem Physiol. 2019;155:101–107. [DOI] [PubMed] [Google Scholar]
- 218. Chen L, Heng J, Qin S, Bian K. A comprehensive understanding of the biocontrol potential of Bacillus velezensis LM2303 against Fusarium head blight. PLoS One. 2018;13:e0198560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Karuppiah V, He A, Lu Z, Wang X, Li Y, Chen J. Trichoderma asperellum GDFS1009‐mediated maize resistance against Fusarium graminearum stalk rot and mycotoxin degradation. Biol Control. 2022;174:105026. [Google Scholar]
- 220. Błaszczyk L, Basińska‐Barczak A, Ćwiek‐Kupczyńska H, Gromadzka K, Popiel D, Stępień Ł. Suppressive effect of Trichoderma spp. on toxigenic Fusarium species. Pol J Microbiol. 2017;66:85–100. [DOI] [PubMed] [Google Scholar]
- 221. Tian Y, Tan Y, Yan Z, Liao Y, Chen J, De Boevre M, et al. Antagonistic and detoxification potentials of Trichoderma isolates for control of zearalenone (ZEN) producing Fusarium graminearum . Front Microbiol. 2018;8:2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Tian Y, Yu D, Liu N, Tang Y, Yan Z, Wu A. Confrontation assays and mycotoxin treatment reveal antagonistic activities of Trichoderma and the fate of Fusarium mycotoxins in microbial interaction. Environ Pollut. 2020;267:115559. [DOI] [PubMed] [Google Scholar]
- 223. Galletti S, Paris R, Cianchetta S. Selected isolates of Trichoderma gamsii induce different pathways of systemic resistance in maize upon Fusarium verticillioides challenge. Microbiol Res. 2020;233:126406. [DOI] [PubMed] [Google Scholar]
- 224. Kemp ND, Vaughan MM, McCormick SP, Brown JA, Bakker MG. Sarocladium zeae is a systemic endophyte of wheat and an effective biocontrol agent against Fusarium head blight. Biol Control. 2020;149:104329. [Google Scholar]
- 225. Li P, Bhattacharjee P, Wang S, Zhang L, Ahmed I, Guo L. Mycoviruses in Fusarium species: an update. Front Cell Infect Microbiol. 2019;9:257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226. Paudel B, Pedersen C, Yen Y, Marzano SYL. Fusarium graminearum virus‐1 strain Fgv1‐SD4 infection eliminates mycotoxin deoxynivalenol synthesis by Fusarium graminearum in FHB. Microorganisms. 2022;10:1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227. Li P, Zhang H, Chen X, Qiu D, Guo L. Molecular characterization of a novel hypovirus from the plant pathogenic fungus Fusarium graminearum . Virology. 2015;481:151–160. [DOI] [PubMed] [Google Scholar]
- 228. Li K, Liu D, Pan X, Yan S, Song J, Liu D, et al. Deoxynivalenol biosynthesis in Fusarium pseudograminearum significantly repressed by a megabirnavirus. Toxins. 2022;14:503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Smaoui S, Agriopoulou S, D'Amore T, Tavares L, Mousavi Khaneghah A. The control of Fusarium growth and decontamination of produced mycotoxins by lactic acid bacteria. Crit Rev Food Sci Nutr. 2022;63:1–28. [DOI] [PubMed] [Google Scholar]
- 230. Nasrollahzadeh A, Mokhtari S, Khomeiri M, Saris PEJ. Antifungal preservation of food by lactic acid bacteria. Foods. 2022;11:395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231. Mirza Alizadeh A, Golzan SA, Mahdavi A, Dakhili S, Torki Z, Hosseini H. Recent advances on the efficacy of essential oils on mycotoxin secretion and their mode of action. Crit Rev Food Sci Nutr. 2022;62:4726–4751. [DOI] [PubMed] [Google Scholar]
- 232. Singh P, Dasgupta N, Singh V, Chandra Mishra N, Singh H, Purohit SD, et al. Inhibitory effect of clove oil nanoemulsion on fumonisin isolated from maize kernels. LWT. 2020;134:110237. [Google Scholar]
- 233. Liu M, Zhao L, Gong G, Zhang L, Shi L, Dai J, et al. Invited review: remediation strategies for mycotoxin control in feed. J Anim Sci Biotechnol. 2022;13:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234. Sovrani V, Blandino M, Scarpino V, Reyneri A, Coïsson JD, Travaglia F, et al. Bioactive compound content, antioxidant activity, deoxynivalenol and heavy metal contamination of pearled wheat fractions. Food Chem. 2012;135:39–46. [Google Scholar]
- 235. Lee US, Lee MY, Park WY, Ueno Y. Decontamination of Fusarium mycotoxins, nivalenol, deoxynivalenol, and zearalenone, in barley by the polishing process. Mycotoxin Res. 1992;8:31–36. [DOI] [PubMed] [Google Scholar]
- 236. Cheli F, Pinotti L, Rossi L, Dell'Orto V. Effect of milling procedures on mycotoxin distribution in wheat fractions: a review. LWT Food Sci Technol. 2013;54:307–314. [Google Scholar]
- 237. Lu X, Naidis GV, Laroussi M, Reuter S, Graves DB, Ostrikov K. Reactive species in non‐equilibrium atmospheric‐pressure plasmas: generation, transport, and biological effects. Phys Rep. 2016;630:1–84. [Google Scholar]
- 238. Yousefi M, Mohammadi MA, Khajavi MZ, Ehsani A, Scholtz V. Application of novel non‐thermal physical technologies to degrade mycotoxins. J Fungi. 2021;7:395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239. Feizollahi E, Iqdiam B, Vasanthan T, Thilakarathna MS, Roopesh MS. Effects of atmospheric‐pressure cold plasma treatment on deoxynivalenol degradation, quality parameters, and germination of barley grains. Appl Sci. 2020;10:3530. [Google Scholar]
- 240. Wielogorska E, Ahmed Y, Meneely J, Graham WG, Elliott CT, Gilmore BF. A holistic study to understand the detoxification of mycotoxins in maize and impact on its molecular integrity using cold atmospheric plasma treatment. Food Chem. 2019;301:125281. [DOI] [PubMed] [Google Scholar]
- 241. FAO . A scheme and training manual on good agricultural practices (GAP) for fruits and vegetables. Rome: FAO; 2016. [Google Scholar]
- 242. Imade F, Ankwasa EM, Geng H, Ullah S, Ahmad T, Wang G, et al. Updates on food and feed mycotoxin contamination and safety in Africa with special reference to Nigeria. Mycology. 2021;12:245–260. [DOI] [PMC free article] [PubMed] [Google Scholar]


