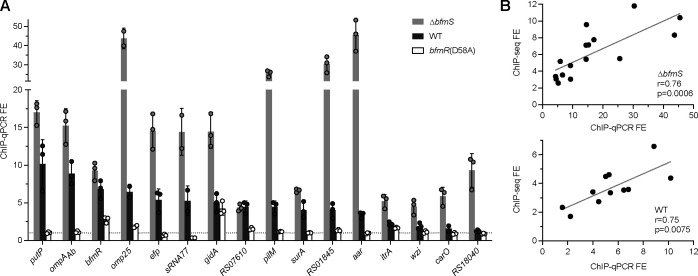
Fig. 6. Independent ChIP-qPCR experiments validate phosphorylation-dependent BfmR binding at select target sites.
(A) Independent ChIP was performed with NRA49 (ΔbfmS), NRA28 (WT), and NRA29 [bfmR(D58A)]. qPCR was used to calculate fold enrichments at binding sites predicted by ChIP-seq. Binding sites are labelled by the closest gene. Dotted line denotes FE value of 1. Data points show mean ± s.d. (n=3; except omp25 and sRNA77, n=2). (B) Correlation of the FE values from ChIP-Seq and independent ChIP-qPCR for NRA49 (top) and NRA28 (bottom). Correlations were evaluated by Pearson’s r. Gray line shows simple linear regression.