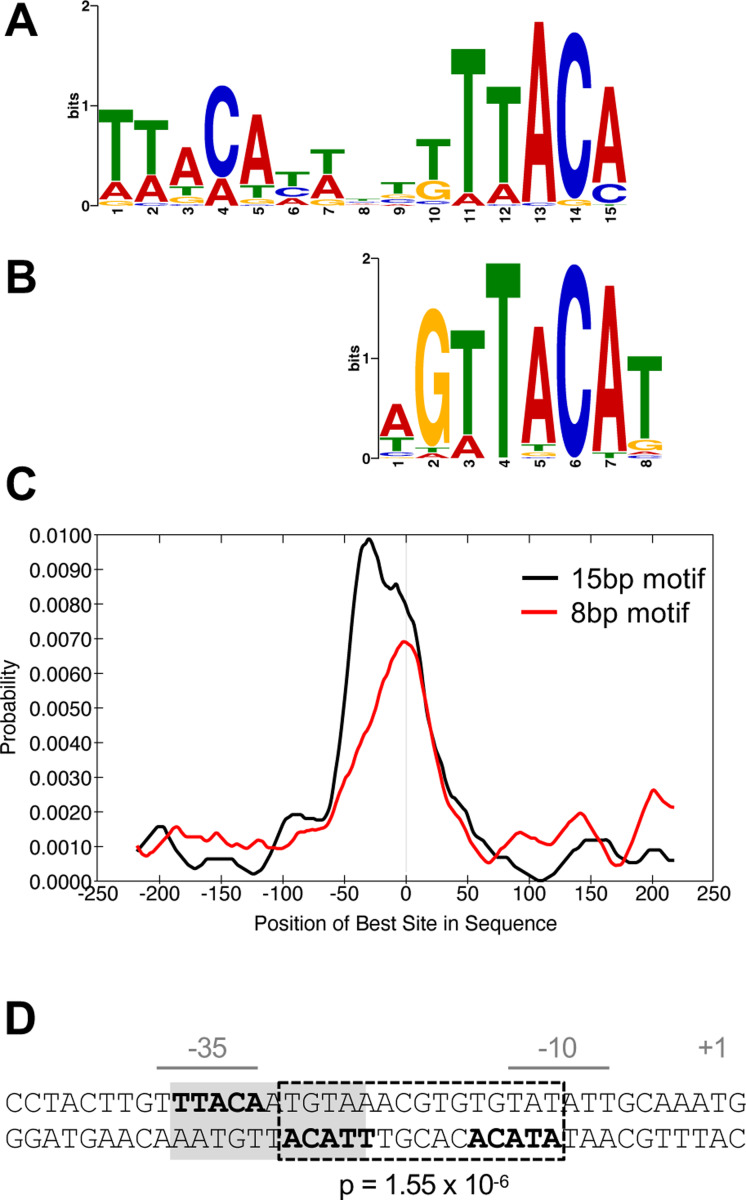
Fig. 7. BfmR-targeted DNA motifs.
(A-C) Identification and distribution of BfmR-binding motifs using a set of 87 input sequences associated with a ChIP-seq peak FE >3 in NRA49 (ΔbfmS). (A) 15bp BfmR-binding motif identified from MEME motif discovery program (E-value 1.3×10−13). (B) 8bp binding motif identified from STREME motif discovery program (E-value 3.9×10−3). (C) Distribution of the best matches to the motif in the binding site sequences as analyzed by using CentriMo. The vertical line in the center of the graph corresponds to the center of the sequences. (D) Location of a BfmR binding motif in the BfmR promoter. The promoter identified in (8) is shown. The repeated half sites are in bold. Dotted box denotes a 15bp direct-repeat BfmR binding motif determined by FIMO (p-value shown). Grey shading denotes the previously identified inverted repeat (23).