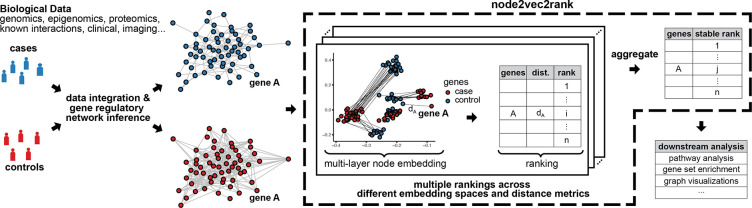
Figure 1:
Illustration of node2vec2rank and an analysis pipeline for a case-control study with two graphs. From the biological data, two gene regulatory networks are computationally inferred and subsequently contrasted using n2v2r to identify their differences. Node2vec2rank uses a multi-layer node embedding algorithm to create two sets of vector representations for all genes (depicted here in two dimensions). For every gene, n2v2r computes the disparity between its two representations, which is then used to rank the genes in descending order of disparities. The process is repeated multiple times, producing different embedding spaces (with dimensionalities of 2, 4, 8, 16, . . . ) and ranking based on different distance metrics (cosine, Euclidean, etc.). The multiple rankings are aggregated to produce a final stable ranking. The output is readily available to be used in downstream applications including visualizing the largest differences and performing gene set enrichment analysis.