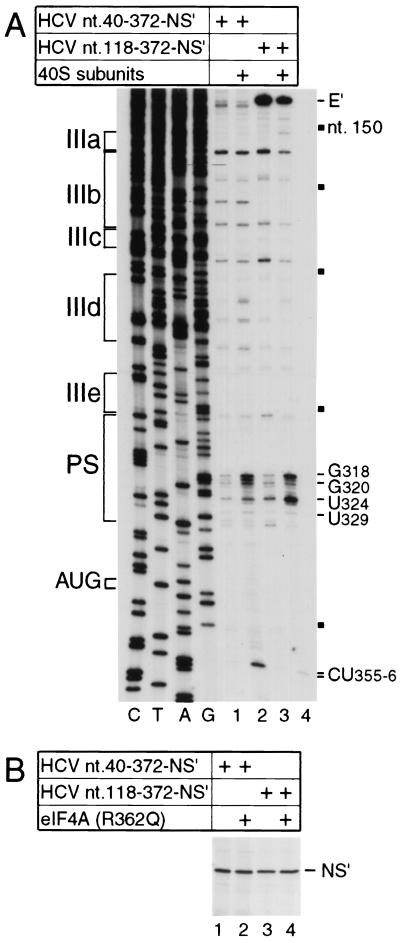
FIG. 2.
Influence of domain II on IRES function. (A) Toeprint analysis of binary ribosomal complex formation on the HCV IRES. Ribosomal 40S subunits were incubated with HCV nt 40-372.NS′ or nt 118-372.NS′ mRNA under standard reaction conditions and then analyzed by primer extension. Full-length nt 118-372.NS′ cDNA is marked E′; other cDNA products terminated at the sites indicated on the right. Reference lanes C, T, A, and G depict HCV sequences; IRES subdomains IIIa, IIIb, IIIc, IIId, IIIe, and PS (pseudoknot) are indicated on the left; HCV nucleotides are indicated by black squares at 50-nt intervals from nt 150 to 350 on the right. (B) Translation in RRL of HCV nt 40-372.NS′ mRNA (lanes 1 and 2) or nt 118-372.NS′ mRNA (lane 3 and 4) in the presence (lanes 2 and 4) or absence (lanes 1 and 3) of a 10-fold molar excess of mutant eIF4A(R362Q). Translation products were analyzed by autoradiography after electrophoresis on a 12% polyacrylamide gel. The position of the NS′ translation product is indicated.