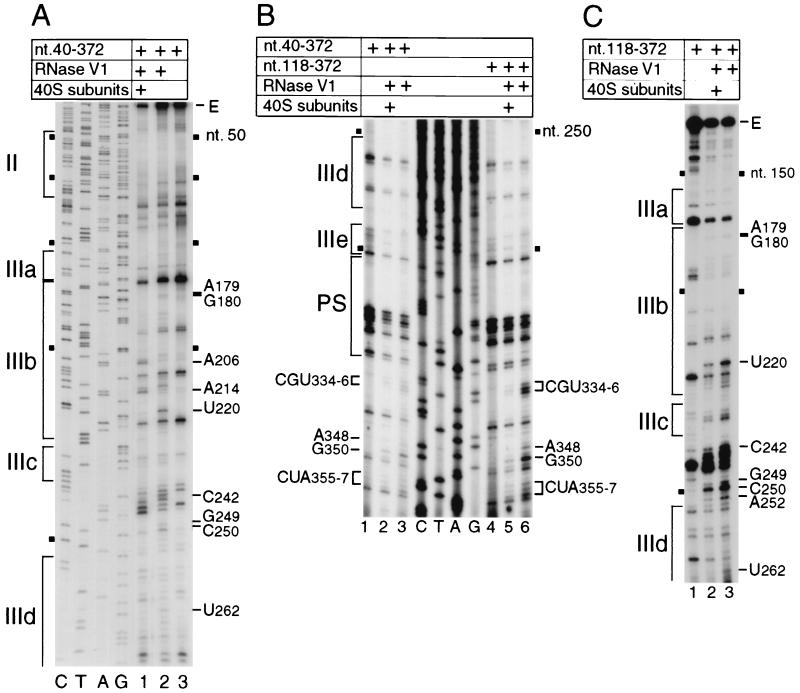
FIG. 3.
RNase V1 footprinting of the binary 40S subunit-HCV IRES complex. The gels show polyacrylamide-urea gel fractionation of cDNA products obtained after primer extension. (A) Sensitivity of HCV nt 40-372 RNA upstream of nt 272 to cleavage (lanes 1 and 2) either alone (lane 2) or bound by a 40S subunit (lane 1); (B) sensitivity of HCV nt 40-372 RNA (lanes 1 to 3) or nt 118-372 RNA (lanes 4 to 6) upstream of nt 360 to cleavage (lanes 2, 3, 5, and 6) either alone (lanes 3 and 6) or bound by a 40S subunit (lanes 2 and 5); (C) sensitivity of HCV nt 118-372 RNA upstream of nt 265 to cleavage (lanes 2 and 3) either alone (lane 3) or bound by a 40S subunit (lane 2). cDNA products obtained after primer extension of untreated HCV RNA are shown in lane 3 of panel A, lanes 1 and 4 of panel B, and lane 1 of panel C. A dideoxynucleotide sequence generated with the same primer (shown in lanes C, T, A, and G in panels A and B) was run in parallel on each gel. IRES subdomains II, IIIa, IIIb, IIIc, IIId, IIIe, and PS (pseudoknot), as appropriate, are indicated on the left of each panel; HCV nucleotides are indicated by black squares at 50-nt intervals, and the positions of protected residues are indicated on the sides of each panel.