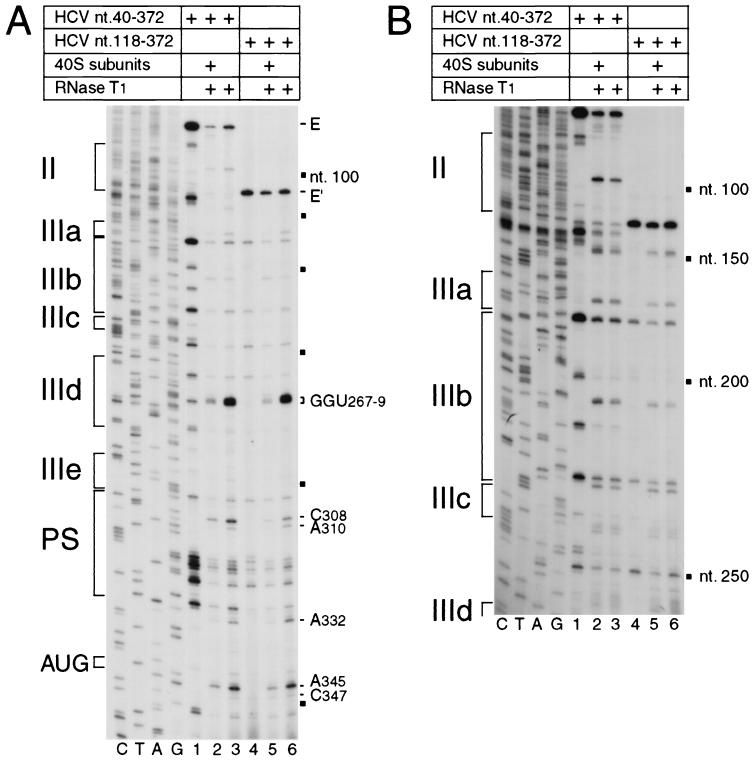
FIG. 4.
RNase T1 footprinting of the binary 40S subunit-HCV IRES complex. Polyacrylamide-urea gel fractionation of cDNA products obtained after primer extension shows the sensitivity of HCV nt 40-372 RNA (lanes 1 to 3) or nt 118-372 RNA (lanes 4 to 6) upstream of nt 360 (A) or of nt 250 (B) to cleavage (lanes 1, 2, 5, and 6) either alone (lanes 3 and 6) or bound by a 40S subunit (lanes 2 and 5). cDNA products obtained after primer extension of untreated HCV RNA are shown in lanes 1 and 4 of both panels. A dideoxynucleotide sequence generated with the same primer and run in parallel is shown to the left of both panels. IRES subdomains II, IIIa, IIIb, IIIc, IIId, IIIe, and PS (pseudoknot) are indicated on the left of each panel; HCV nucleotides are indicated by black squares at 50-nt intervals, and the positions of protected residues are marked on the right of each panel. The position of the initiation codon AUG is indicated to the left of panel A.