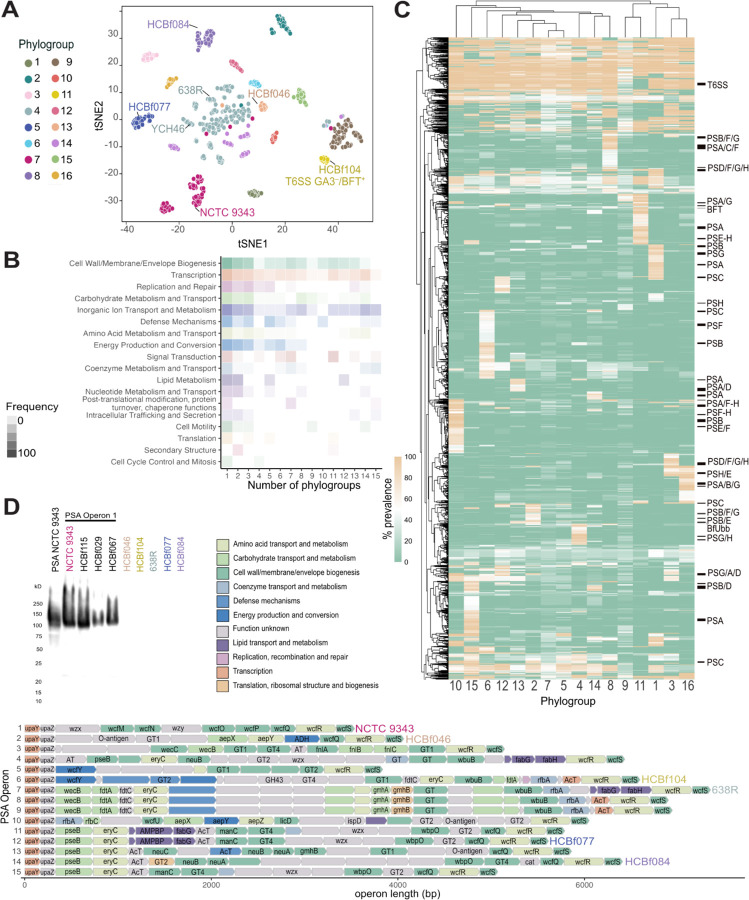
Figure 2. B. fragilis phylogroups have unique core-phylogroup genes.
A) B. fragilis phylogroups based on t-distributed Stochastic Neighbor Embedding (t-SNE) algorithm of a MASH, k-mer based distance matrix of the whole genome sequence of all strains. Phylogroups labeled 1–16. The commonly used laboratory strains, NCTC 9343, YCH46, and 638R, are labeled within their respective phylogroups along with other B. fragilis strains used in functional assays described in this study: HCBf046, HCBf084, HCBf077, and HCBf104. T6SS GA3–/BFT+ represents a cluster of strains that lack T6SS GA3 and are positive for the presence of bft.
B) Proportion of genes that are core to a collection of phylogroups, between 1 and 15, divided into COG categories with proportion expressed by the alpha of the heatmap cells.
C) A heatmap of gene prevalence in each phylogroup. A euclidean clustering algorithm was applied to the rows and columns, gene clusters that belong to capsular polysaccharide paths are labeled, as well as the T6SS GA3 gene cluster and the location of bft and BfUbb.
D) Western blot analysis of PSA of B. fragilis strains representing six PSA operon structures. Rabbit anti-PSA antibody was raised against NCTC 9343, which representing PSA operon 1 (top left). PSA operon structure of B. fragilis strains of high-quality assemblies (n=262). Genes colored by COG category and annotated with a gene name, if available by Bakta annotation.