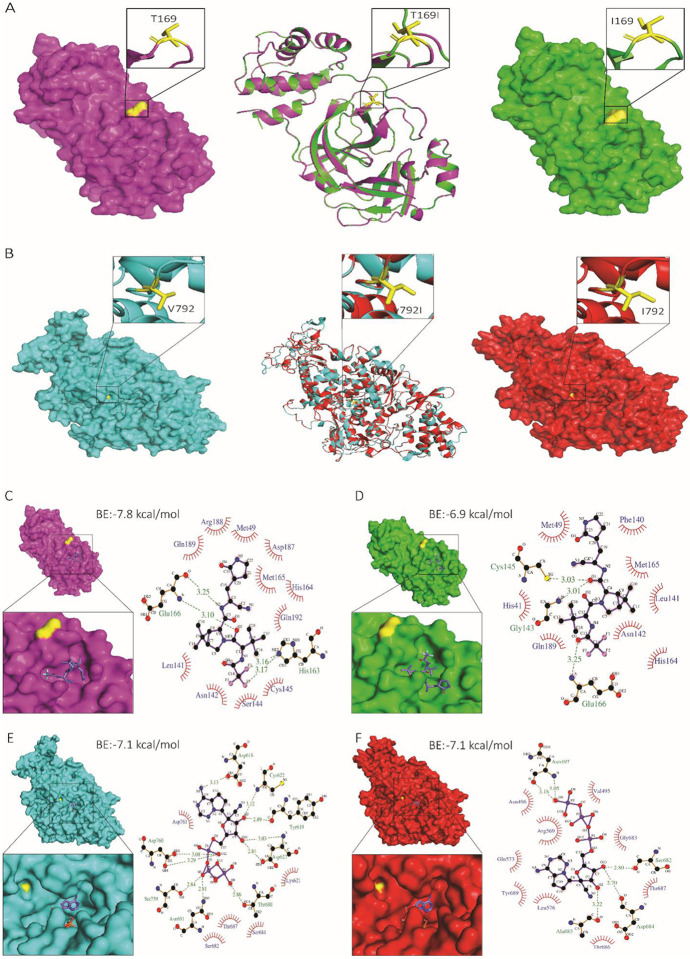
Fig. 4: Structural analysis of nsp5 T169I and nsp12 V792I substitutions.
Comparison of nsp5 (A) and nsp12 (B) protein models between WT (NYI45-21) and mutant nsp5T169I and nsp12V792I proteins. (A, B, left panel) WT: Surface representation of nsp5 (magenta) and nsp12 (cyan) proteins with specific amino acid substitutions highlighted in the inset for each protein. The position of substituted amino acids is depicted in yellow sticks with proper labelling. (A, B, middle panel) Superimposed Image: Cartoon representation of the superimposed image of WT (magenta/cyan) and nsp5T169I and nsp12V792I proteins (green/red), depicting specific substitutions in the inset box. (A, B, right panel) nsp5T169I and nsp12V792I: nsp5 (magenta) and nsp12 (cyan) proteins with specific amino acid substitutions highlighted in inset for each protein in box. The position of the substituted amino acids is shown in yellow sticks with proper labelling. Note: The protein models of the nsp5T169I and nsp12V792I proteins was generated using Swiss MODELLER, resulting in RMSD values of 0.003 Å for nsp5 and 0.026 Å for nsp12 proteins, indicating high structural similarity. Surface representation of nsp5 proteins of (C) WT and (D) nsp5T169I protein docked to nirmatrelvir drug molecules shown in the inset using PyMOL analysis tool. 2D depiction of nsp5 protein residues interactions with nirmatrelvir drug molecules via LIGPLOT+ tool where hydrogen bonds are showed in green dotted lines with a distance in Ångström (Å) and hydrophobic interactions are showed in curvature. Surface representation of nsp12 proteins of (E) WT and (F) mutant nsp12V792I docked remdesivir drug molecules shown in inset for each using PyMOL tool. 2D depiction of nsp12 protein residues interactions with remdesivir drug molecule via LIGPLOT+ tool where hydrogen bond showed in green dotted lines with a distance in Å and hydrophobic interactions are showed in curvature. The colour scheme is consistent throughout the figures, with magenta representing nsp5 and cyan representing nsp12 of WT isolate, and green representing nsp5 and red representing nsp12 of nsp5T169Insp12V792I isolate with substituted amino acid highlighted in yellow. Note: BE: Binding energy.