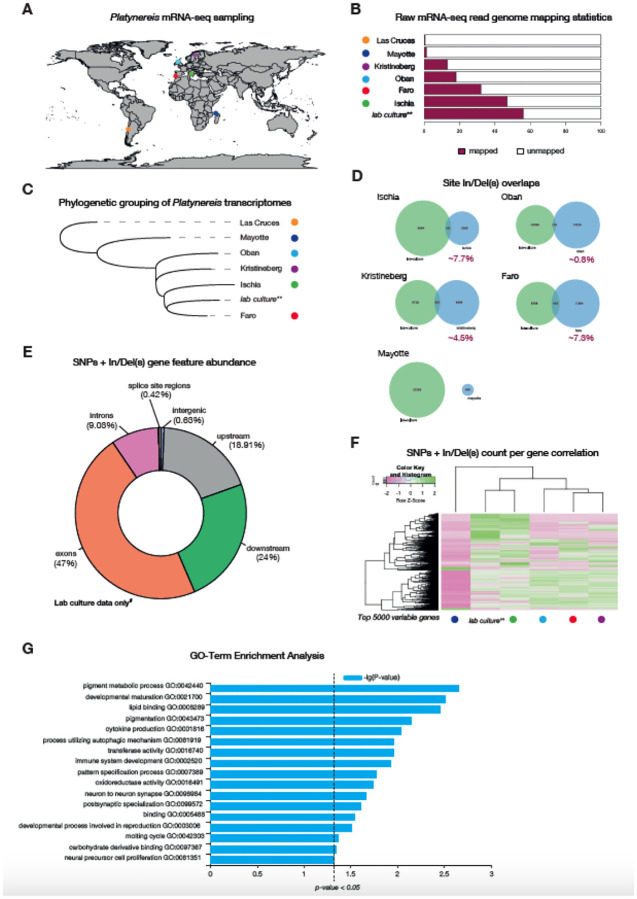
Figure 5. Genomic and transcriptomic variation analyses on wild sampled P. dumerilii.
A, global map of sites of P. dumerilii mRNA sampling. B, histogram of raw mRNA-seq genome mapping percentages. C, phylogenetic grouping/sorting of wild sampled Platynereis transcriptomes via OrthoFinder. D, proportion of In/Del overlaps identified from the different Platynereis samples. E, gene feature abundance/occupancy of SNPs and In/Dels from mRNA-seq reads accessed from P. dumerilii lab cultures. F, SNP and In/Del counts from the same position on the genome correlation for the top 5,000 most variable genes (i.e. genes that showed the most variation in SNP and In/Del counts across the different sites). G, GO-term enrichment analysis of the top 5,000 variable genes.