Figure 1.
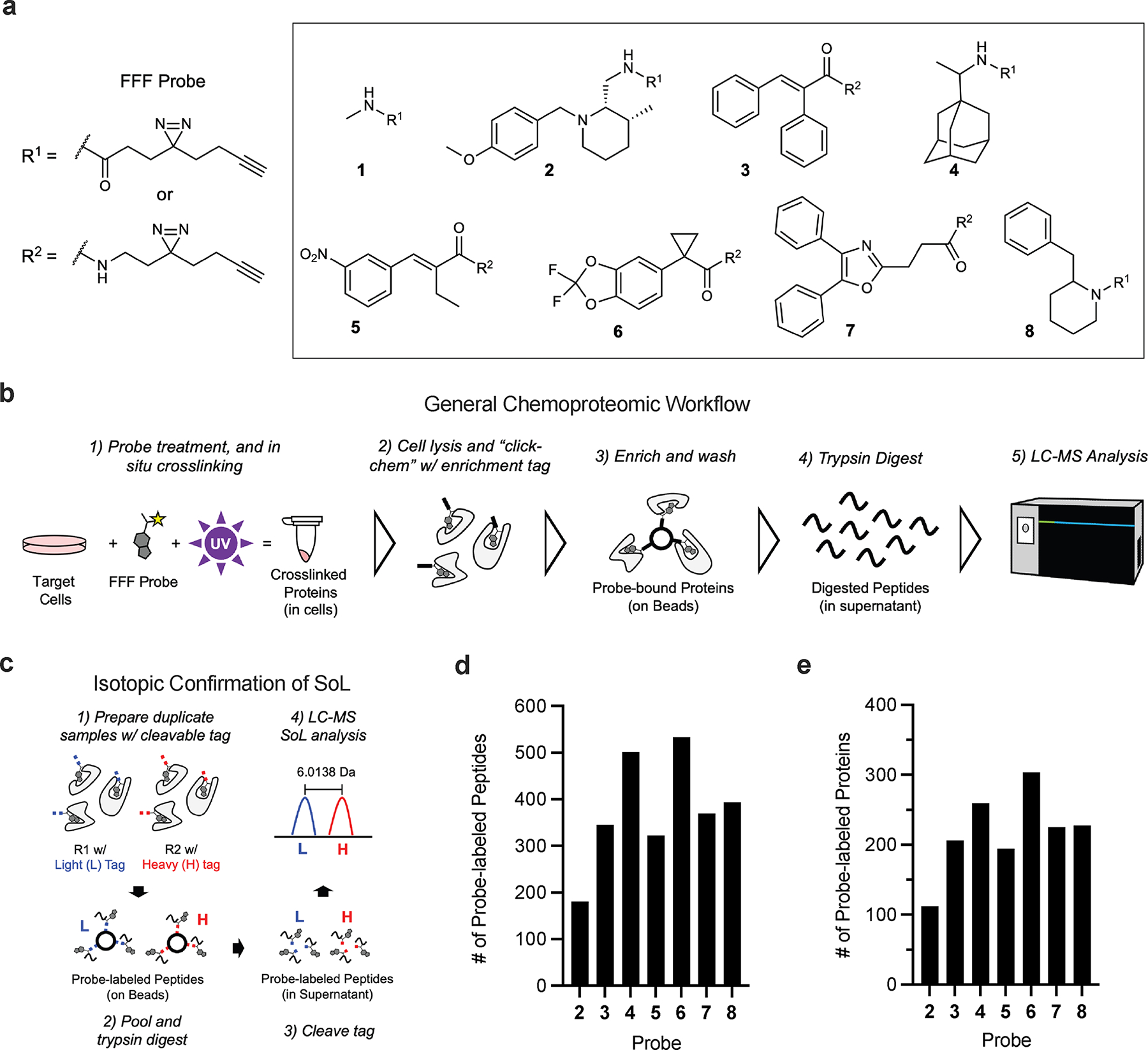
Overview and benchmarking of chemoproteomic site of labeling (SoL) analysis of photoaffinity probes. (a) Structures of fully functionalized fragment (FFF) probes. (b) Schematic of general chemoproteomic workflow and (c) of isotopic confirmation of photoaffinity probe SoL. Briefly, cells are treated with an FFF probe before being crosslinked under UV light. Cells are then lysed and a biotin enrichment tag is appended to crosslinked proteins via copper-catalyzed azide-alkyne cycloaddition (CuAAC) “click” chemistry. Following the click reaction, captured proteins are enriched via streptavidin beads and digested with trypsin. Digested peptides are then labeled with TMT reagents and pooled for multiplexed, quantitative proteomic analysis. Reporter ion signals for proteins in FFF probe-treated samples are compared to the methyl probe-treated samples to determine relative enrichment levels at the total protein level. Probe-labeled peptides (d) and proteins (e) detected in benchmarking SoL workflow.
