Figure 3.
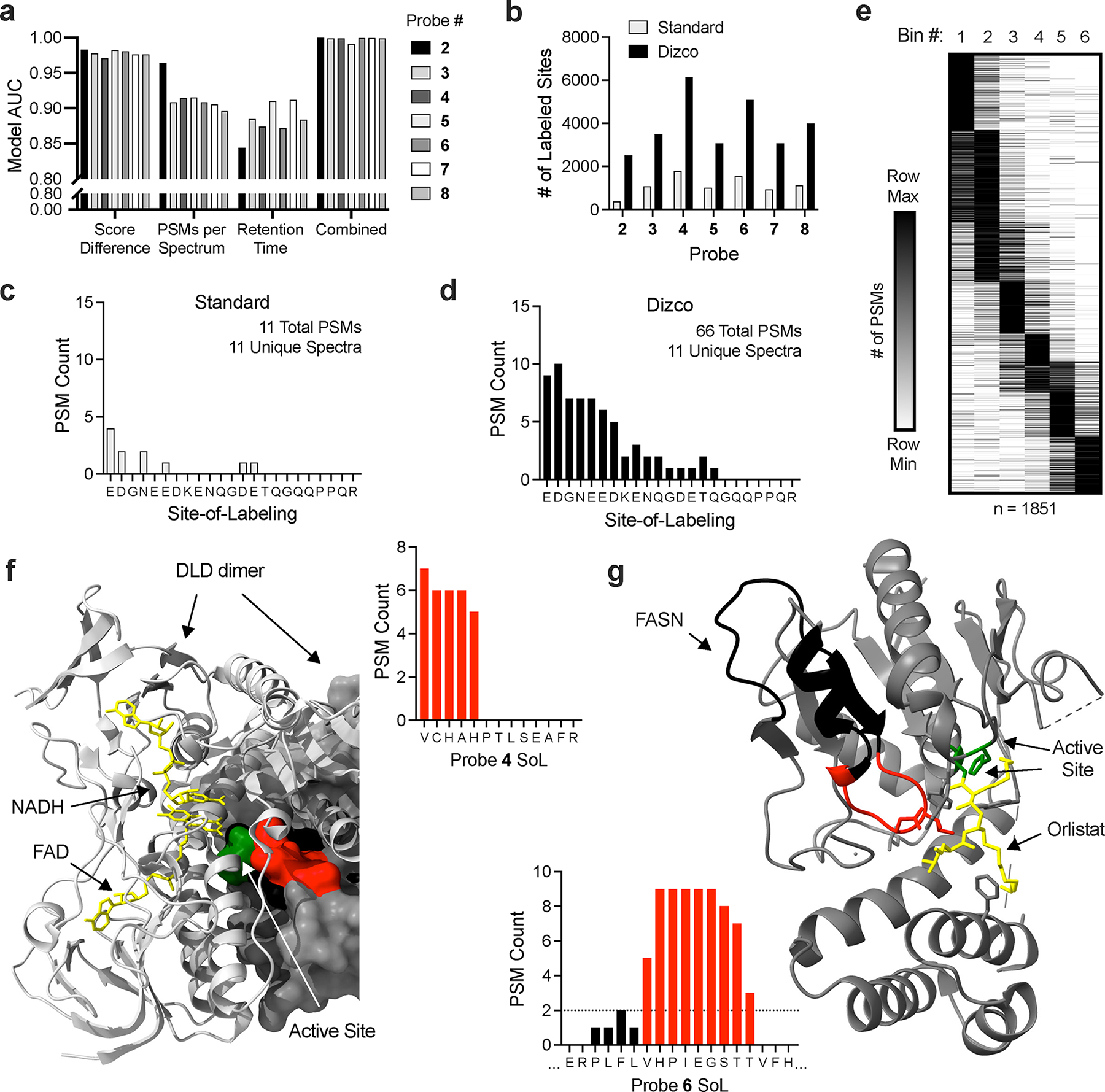
Dizco workflow increases depth of SoL experiments and confidence of label locations. (a) Predictive capabilities of individual model features and combined final models for each probe. (b) Number of labeled sites detected in standard and Dizco workflows. (c-d) Example of a peptide label distribution detected by the standard (c) and Dizco (d) workflows. (e) Clustered heatmap of peptide label locations from the Dizco workflow binned into six bins across peptide sequences. (f-g) Examples of high-confidence sites detected in the Dizco workflow. For all structures, labeled peptide residues are colored red and the remainder of each peptide is colored black. Active/other indicated sites are colored green and co-resolved ligands are colored yellow. (f) DLD probe label site overlapping with active and ligand binding sites (PDB: 1ZMD). (g) FASN probe label site overlapping with active site and inhibitor binding sites (PDB: 2PX6).
