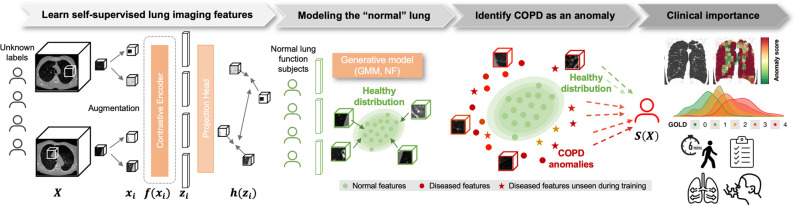
Fig. 2.
Overview of the pipeline. Preprocessed 3D patches xi are extracted from the lung parenchyma from Insp and ExpR CT images X. Informative lung representations zi at the patch level are learned through a self-supervised contrastive pretext task f(xi), based on the rationale of clustering together unlabeled positive pairs while pushing apart negative pairs. A generative model based on a Gaussian mixture model (GMM) or normalizing flow (NF) is then applied on the representations from normal regions of “control” individuals, so that the normal patient lung can be modeled. From this healthy distribution, anomalies/deviations are then detected as COPD-like regions. An anomaly score is assigned per patch based on the negative log likelihood. Patient-level predictions S(X) are obtained by aggregating the anomaly scores from all patches of the lung. Clinical meaning of these scores can be derived with anomaly maps. Further, patient score distribution can be correlated to severity stages and pulmonary function tests, as the St. George’s Respiratory Questionnaire, 6-min walking test, modified Medical Research Council dyspnea score, and spirometry