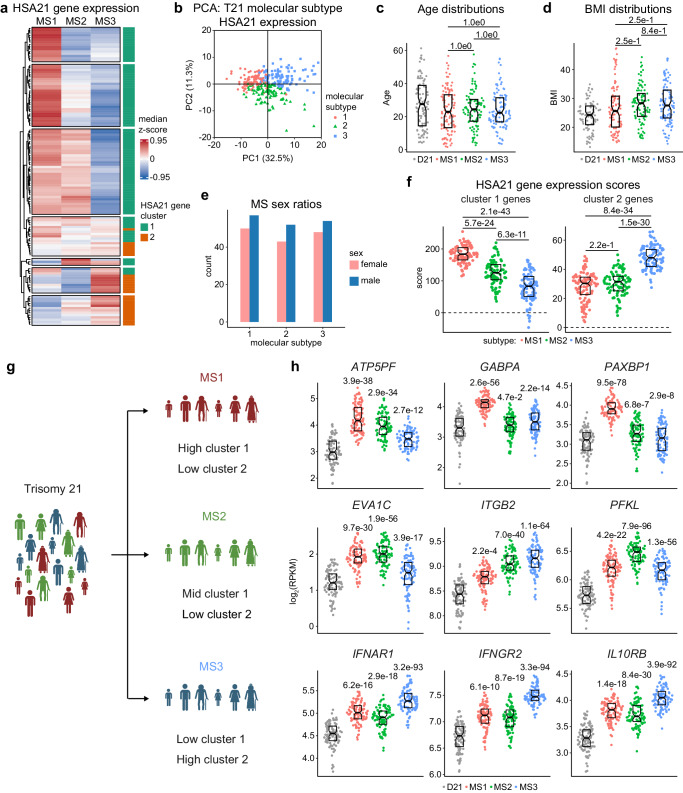
Fig. 2. Variable chromosome 21 gene expression distinguishes molecular subtypes in Down syndrome.
a Heatmap showing unsupervised hierarchical clustering of HSA21 gene expression in trisomy 21 (T21) molecular subtype 1 (MS1) (n = 107), MS2 (n = 95) and MS3 (n = 102). Values are median z-scores across all T21 (n = 304). Colors indicate HSA21 gene cluster 1 (teal) and cluster 2 (orange). b Principal component analysis showing separation of subtypes based on HSA21 gene expression. c, d Sina plots showing similar age (c) and BMI (d) distributions between euploid controls (D21, n = 96, gray), MS1 (n = 107, red), MS2 (n = 95, green), and MS3 (n = 102, blue), with no significant differences as determined by two-sided Wilcoxon rank-sum tests. Statistics represent adjusted p-values (q-values) after Benjamini–Hochberg adjustment for multiple hypotheses. Boxes represent interquartile ranges and medians, with notches approximating 95% confidence intervals. e Bar charts showing similar sex ratios across subtypes. Colors indicate females (pink) and males (blue). f Sina plots showing HSA21 gene cluster polygenic scores in MS1 (n = 107, red), MS2 (n = 95, green), and MS3 (n = 102, blue). Scores were derived for each HSA21 gene cluster by calculating the sum of their z-scores relative to D21 (n = 96). Dashed line indicates D21 mean values. Boxes represent interquartile ranges and medians, with notches approximating 95% confidence intervals. Statistics indicate q-values from linear regressions, adjusted using the Benjamini–Hochberg method. g Illustration showing distinct molecular subtypes among individuals with T21, differentiated by unique overexpression profiles for HSA21 cluster 1 and 2 genes. h Sina plots showing expression of example HSA21 genes in D21 (n = 96, gray), MS1 (n = 107, red), MS2 (n = 95, green), and MS3 (n = 102, blue). Statistics above datapoint swarms are q-values, derived from DESeq2 comparisons to D21 and adjusted using the Benjamini–Hochberg method. Boxes represent interquartile ranges and medians, with notches approximating 95% confidence intervals. Panel g created with BioRender.com released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license.