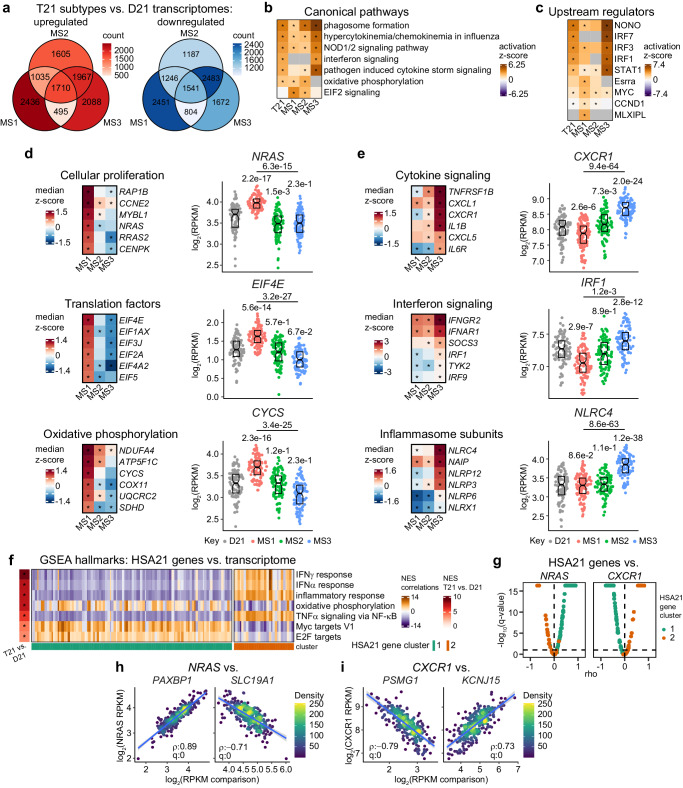
Fig. 3. Molecular subtypes of Down syndrome are distinguished by distinct transcriptomic landscapes.
a Overlapping differentially expressed genes identified by DESeq2 in comparisons of each molecular subtype (MS1, n = 107; MS2, n = 95; MS3, n = 102) against euploid controls (D21, n = 96). b, c Heatmaps showing select canonical pathways (b) and upstream regulators (c) from Ingenuity Pathway Analysis (IPA) of DESeq2 results comparing trisomy 21 (T21, n = 304), MS1 (n = 107), MS2 (n = 95) and MS3 (n = 102) to D21 (n = 96). Asterisks denote q < 0.1 from IPA overrepresentation analysis. d, e Heatmaps display median z-scores for representative genes in MS1 (n = 107), MS2 (n = 95), and MS3 (n = 102) calculated relative to D21 (n = 96), with asterisks denoting q < 0.1 from DESeq2 after Benjamini–Hochberg adjustment. Sina plots illustrate gene expression across groups. q-values, derived from DESeq2 and adjusted using the Benjamini–Hochberg method, are displayed above individual data swarms for comparisons to D21 and above lines for MS3 vs. MS1. Boxes represent interquartile ranges and medians, with notches approximating 95% confidence intervals. f Heatmap depicts pathway enrichment (NES, normalized enrichment score) from Gene Set Enrichment Analysis (GSEA) for HSA21 genes, based on their correlations against the transcriptome in individuals with T21 (n = 304). Left tile annotation shows results from GSEA of DESeq2 comparing T21 (n = 304) vs. D21 (n = 96). Asterisks denote q < 0.1 after Benjamini–Hochberg adjustment. Colors indicate HSA21 gene cluster 1 (teal) and cluster 2 (orange). g Volcano plots showing Spearman correlations of NRAS (left) and CXCR1 (right) vs. HSA21 cluster 1 (teal) and cluster 2 (orange) genes in individuals with T21 (n = 304). Dashed line indicates q = 0.1. h Scatter plots showing expression of NRAS vs. PAXBP1 (HSA21 gene cluster 1) and SLC19A1 (HSA21 gene cluster 2) in individuals with T21 (n = 304). i Scatter plots showing expression of CXCR1 versus PSMG1 (HSA21 gene cluster 1) and KCNJ15 (HSA21 gene cluster 2) in individuals with T21 (n = 304). For (h, i) rho and q-values (Benjamini–Hochberg adjusted p-values) for Spearman correlation are denoted. Points are colored by density; blue lines represent the fitted values from linear regressions, with 95% confidence intervals in grey.