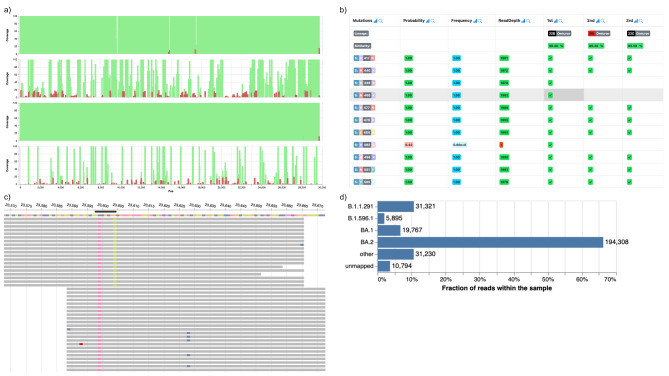
Fig. 2.
Four different example elements of the results generated by UnCoVar: (a) The genome coverage of the aligned reads, visualized for multiple samples, (b) evaluation of known protein alterations from VOCs for one sample, (c) a pileup of reads at the position of one protein alteration. The mutations observed for multiple reads (gray bars) for a single sample, here in the S gene, (d) The lineage assignments inferred for single reads for one sample. Unmapped reads can be attributed to low sequence quality and variation beyond the considered lineages