Figure 6.
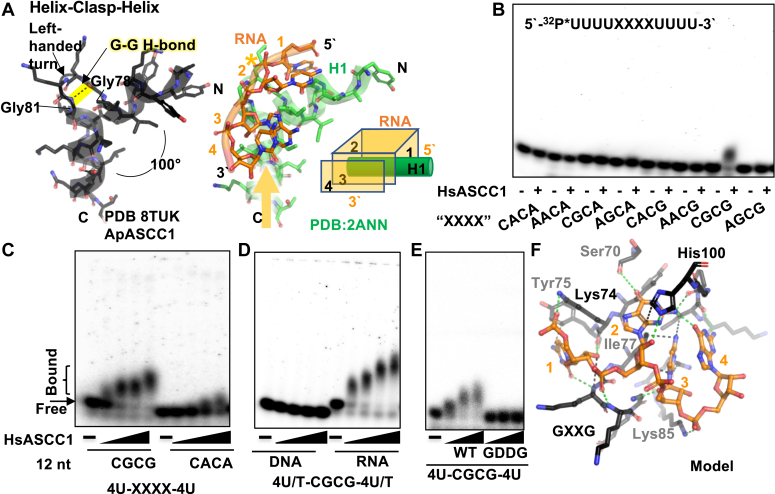
KH Domain Helix-Clasp-Helix structural motif for nucleotide binding is distinct from other two helix nucleic acid interaction motifs.A, structural motif, Helix-Clasp-Helix, found in KH domain of ASCC1 or in representative Nova-1 KH domain bound to RNA (PDB:2ANN). The GXXG motif is positioned by the two helices. In Nova-1, the amino-terminal dipole (yellow arrow) of the second helix is directed towards the RNA phosphate backbone (∗). B, biochemical analysis showing HsASCC1 bound specifically to CGCG-containing RNA. EMSA of 116 nM HsASCC1 incubated with 10 nM radiolabeled 12 nt UUUUXXXXUUUU RNA with XXXX representing specific sequence. C, HsASCC1 sequence specificity for CGCG. Dose-dependent EMSA with varying ASCC1 concentration (60, 120, 240, 360 nM) and 10 nM radiolabeled RNA containing CGCG or CACA. Quantitation in Fig. S4B. D, HsASCC1 sequence specificity for RNA. Dose-dependent EMSA with varying ASCC1 concentration (60, 120, 240, 360 nM) and 10 nM radiolabeled RNA or DNA containing CGCG. Quantitation in Fig. S4C. E, HsASCC1 RNA-binding requires KH domain GXXG motif. Dose-dependent EMSA with varying WT or mutant (GDDG) ASCC1 concentration (60, 120, 240, 360 nM), 10 nM radiolabeled RNA containing CGCG. Quantitation in Fig. S4D. F, Homology-based and energy minimized computational model of ApASCC1 bound to CGCG RNA. Major interactions are colored green (H-bonds, polar) or black (van der Waals). Orientation similar to Figure 5A. Sidechains interacting with RNA are labeled.