FIGURE 5.
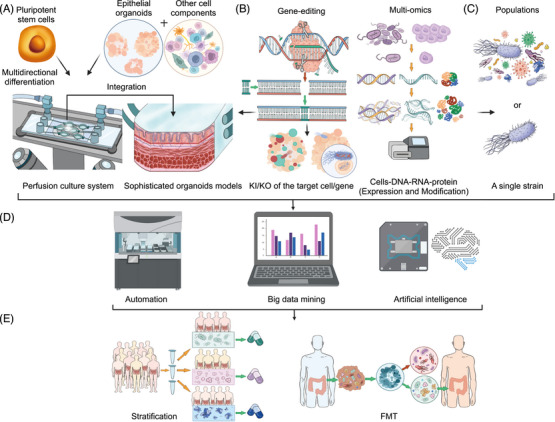
Future trends of microbial research on gut organoids. (A) More sophisticated organoids models, which more closely resemble the structure and function of the gut, can be developed in two ways, either by multipotent stem cell differentiation, or by integrating a variety of immune cells, mesenchymal cells, endothelial cells, and glial cells isolated from the gut. The resulting organoid cultured in a 3D‐printed perfusion system to monitor changes in culture factor and oxygen in real time. (B) The application of omics approaches in studying microbe–host interactions. Biological changes in strain‐host cell interactions have been studied to find and verify the protein targets of the interaction between microbiota and intestinal cells through multiomics and gene‐editing technology. This can be reached by knockout (KO) of the host or bacterial interaction proteins to verify the form of interaction between them, or by knockin (KI) of the target cell/gene into the green fluorescent protein, the strain/specific gene into the red fluorescent protein, or vice versa. Through this way, we can observe the specific interaction between them and analyze the physiological and functional changes (epigenetics, transcriptomics, proteomics, metabolomics, and single cell) of the two sides during the process of interaction. (C) Modeling the effects of the populations (a particular community) or single strains in organoids. (D) Through the automatic collection of organoid model flux tests combined with bioinformatics for big data mining and association analysis, artificial intelligence can be used for treatment plan prediction before biomedical technology used to verify the predicted data. (E) Therapies that target the microbiota and stratify the risk factors for patients with microbial infection, and identify beneficial bacteria for fecal microbiota transplantation (FMT).
