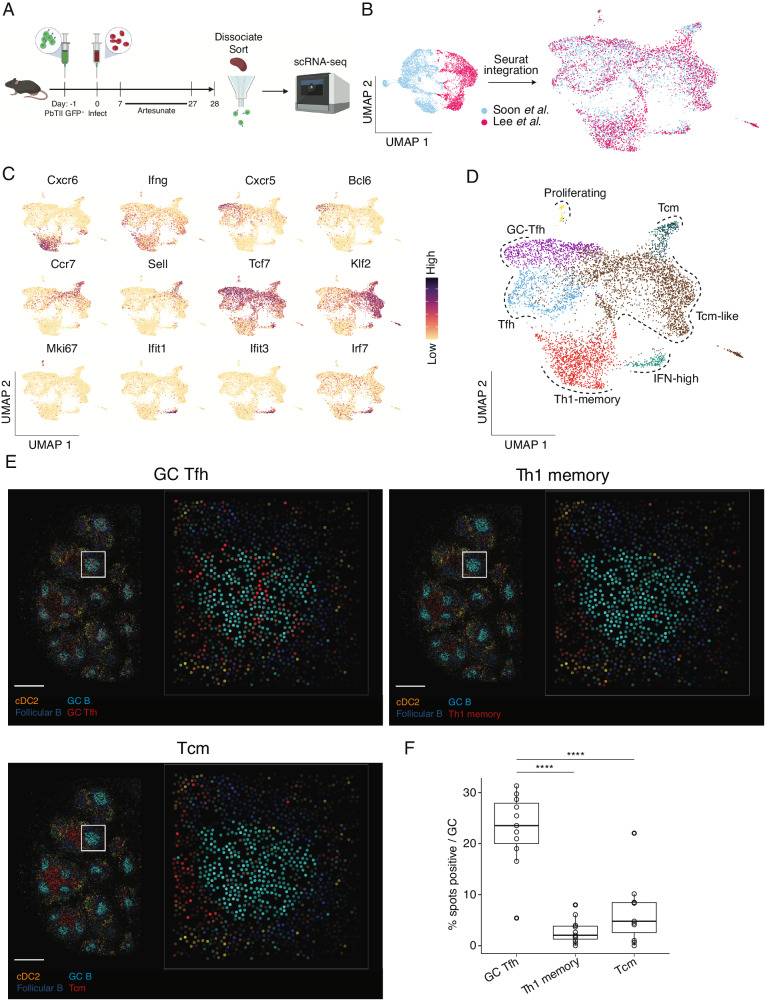
Fig. 1. PbTII cells exhibit a spectrum of transcriptional states in the spleen after primary infection and antimalarial drug treatment.
A Schematic of scRNA-seq experiment to study PbTII cells at day 28 post-infection. B UMAPs of PbTII cell-derived scRNA-seq data before and after integration of our two independent experiments, this study and Soon et al.23—cells coloured by experimental origin. C UMAPs of PbTII cell gene expression patterns for Th1 (Cxcr6, Ifng), Tfh (Cxcr5, Bcl6), TCM (Ccr7, Sell, Tcf7, Klf2), proliferation (Mki67), and IFN-high (Ifit1, Ifit3, Irf7) genes. D UMAP of PbTII cell states depicted by colours and dotted boundaries. E Estimated cell-type abundance for GC Tfh, TCM, and Th1-memory (red), compared with the same cDC2, GC B cells and follicular B cells in each panel, assessed across a Slide-seqV2 spatial transcriptomic map of mouse spleen at day 30 p.i.—scale bar: 500 µm: white boxes shown at higher magnification within each inset panel, with each dot representing a 10 µm bead. F Box plots showing % positive occurrence of GC Tfh, TCM, or Th1-memory cells in each GC (each open circle denoting 1 GC). Cell abundance (computed using cell2location) >0.2 is considered a positive occurrence. Centre line indicates median and box limits upper and lower quartiles; two-sided Wilcoxon signed rank test: ****P < 0.0002 (exact values: P = 0.00019 for GC Tfh vs. Tcm; P = 1.134 × 10−5 for GC Tfh vs. Th1-memory).