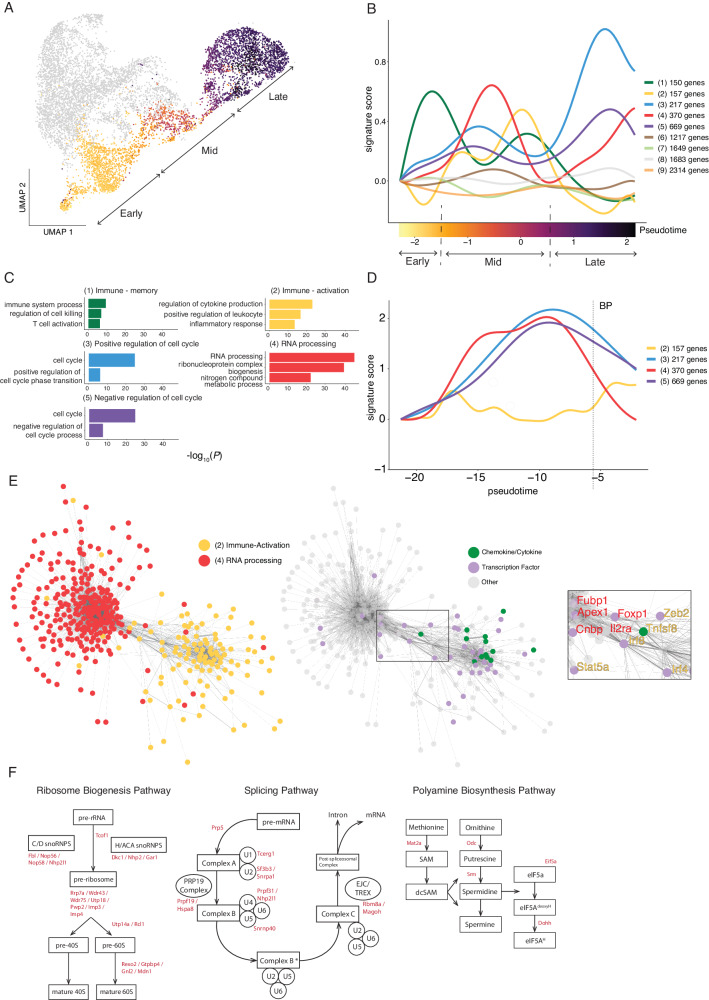
Fig. 4. Transcriptome dynamics of Th1-recall predicts suggest early RNA processing associated with rapid effector function and subsequent proliferation.
A UMAP of antigen-experienced PbTII cells prior to and 1, 2, and 3 days post-re-infection, with only Th1-like cells coloured according to inferred pseudotime values (from 1-dimensional BGPLVM, GPfates), split into early, mid and late-pseudotime. B Expression dynamics for pseudo-temporally variable genes in Th1-like cells along pseudotime, genes grouped according to similar dynamics, represented as signature scores. C Summary of gene ontology enrichment analysis of biological processes associated with genes for selected groups from (B). X-axis represents negative log-transformed P-values that indicate the extent of enrichment of biological processes; Fisher’s exact test is used to identify over-represented GO terms. D Expression dynamics for gene groups 2–5 for Th1-like PbTII cells during primary infection23; BP denotes Th1/Tfh bifurcation point. E Co-expression network analysis of genes (represented as nodes) in Dynamics 2 and 4. Edge weight corresponds to Spearman’s rho values (only showing rho > 0.2). Gene labels in the inset coloured according to the dynamic of origin. F Schematics of enriched biological pathways associated with genes from Dynamic 4. Genes found in Dynamic 4 are highlighted in red.