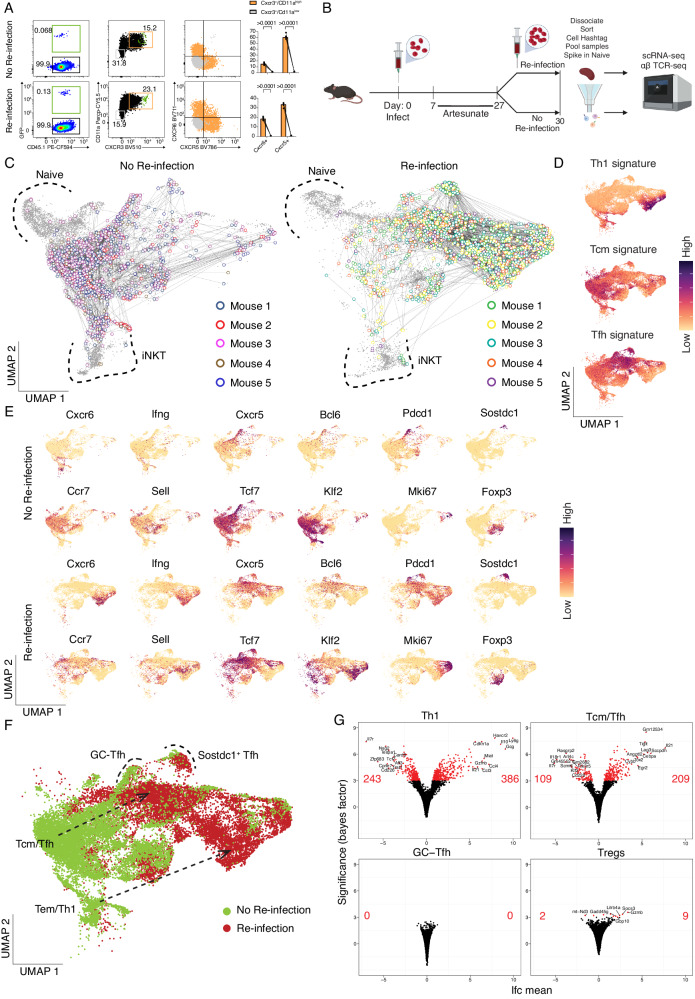
Fig. 6. TCR-diverse, antigen-experienced CD4+ T cells exhibit varied recall responses during re-infection.
A Representative FACS plot showing GFP+ PbTII cells, CD11a and CXCR3 expression, and CXCR6 and CXCR5 expression (orange: gated on CD11ahiCXCR3+, grey: gated on CD11aloCXCR3−) in CD4+ T cells prior to and 3 days after re-infection. Graphs show the percentage of CXCR6+ and CXCR5+ cells from CD11ahiCXCR3+ versus CD11aloCXCR3− CD4+ T cells (n = 5 mice). Statistical analysis was performed using a Wilcoxon signed-ranks test. ****P < 0.0001. B Schematic of scRNA-seq and TCR-seq experiment to study CD11ahiCXCR3+ polyclonal CD4+ T cells prior to and 3 days after re-infection. C UMAP of CD11ahiCXCR3+ and inferred naïve (CD11aloCXCR3-) polyclonal CD4+ T cells. Cells sharing the same TCR chains connected with edges. Only families with four or more cells sharing the same TCR chains are shown in the plot. Naïve cells and iNKT cells are marked with dotted boundaries. D UMAP of CD11ahiCXCR3+ cells expressing Th1, Tcm, and Tfh signature scores. E UMAP of CD11ahiCXCR3+ cells expressing genes associated with Th1 (Cxcr6, Ifng), Tfh (Cxcr5, Bcl6, Pdcd1), TCM (Ccr7, Sell, Tcf7, Klf2), proliferation (Mki67), Sostdc1+ cells, and Treg (Foxp3) cells. F UMAP of CD11ahiCXCR3+ polyclonal CD4+ T cells prior to and 3 days post-re-infection. Differentiation trajectories are indicated with arrows. GC Tfh cells and Sostdc1+ Tfh cells are marked with dotted boundaries. G Volcano plots showing the number of differentially expressed genes (genes with Bayes factor > 3) comparing prior to and after re-infection for each inferred state: Th1 cells (Top-Left), TCM/Tfh cells (Top-Right), GC Tfh cells (Bottom-Left), and Treg cells (Bottom-Right). Number of significantly upregulated/downregulated genes and the top 10 upregulated/downregulated genes annotated on volcano plots.