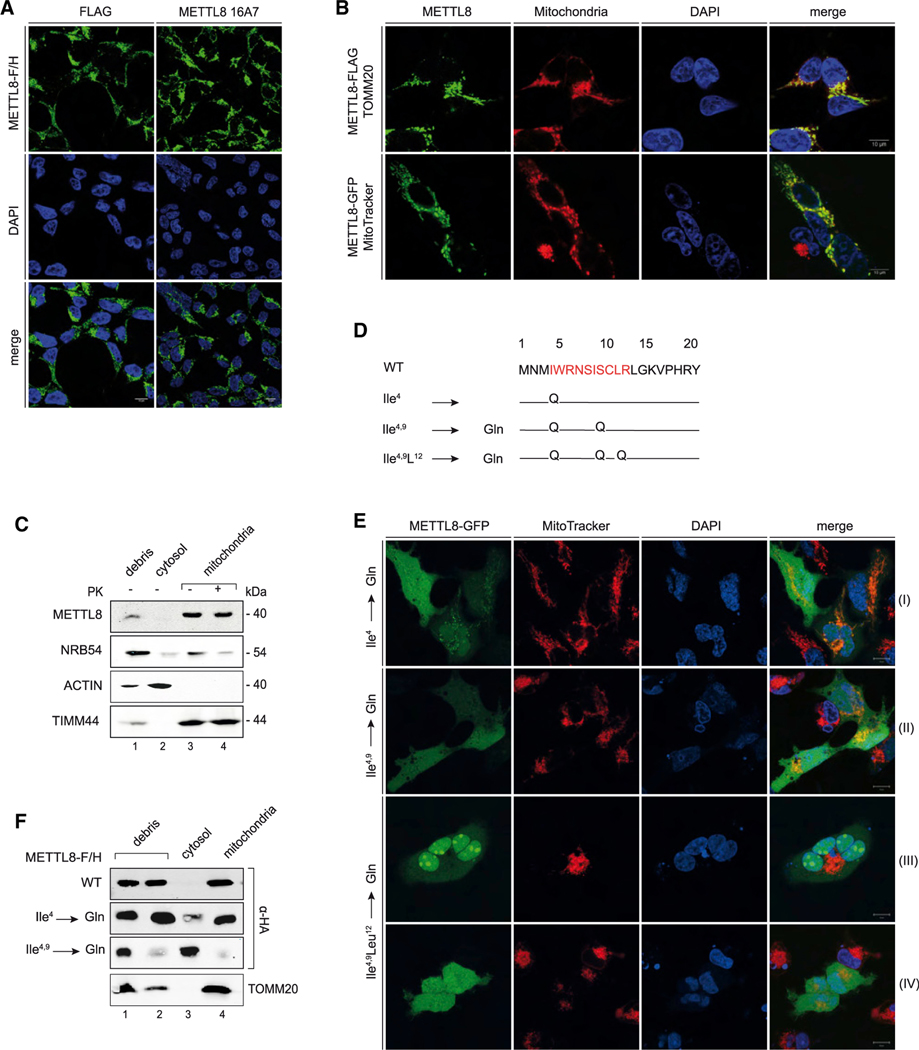
Figure 1. METTL8 is a mitochondrial protein.
(A) Immunofluorescence detection of METTL8-F/H by anti-FLAG (green, left panel) and anti-METTL8 16A7 (green, right panel) in Flp-In TREx293 cells. Nuclei were stained with DAPI (blue) (scale bars: 10 μm).
(B) Subcellular localization of METTL8-F/H (green, top panel) and METTL8-GFP (green, bottom panel) in Flp-In TREx293 cells. Top: immunofluorescence detection of METTL8-F/H (green), mitochondrial import receptor subunit TOMM20 (red), and DAPI (blue). Bottom: fluorescence imaging of METTL8-GFP (green), MitoTracker Deep Red (red), and Hoechst 33342 (blue) (scale bars: 10 μm).
(C) Fractionation of Flp-In TREx293 cell lysates into debris/nuclei (NRB54), cytosol (ACTIN) and mitochondria (TIMM44). Mitochondria were treated with Proteinase K. Endogenous METTL8 was detected after anti-METTL8 IP.
(D) Snapshot of the predicted MTS pre-sequence with the MPP (mitochondrial-processing peptidase) cleavage site (position 20) and mutation of amino acids Ile4 and 9 and Leu12 to glutamines.
(E) Live cell imaging of modified METTL8-GFP (green) by fluorescence detection of MitoTracker Deep Red (red) and Hoechst 33342 (blue). Fluorescence detection of the METTL8-GFP single (row I), double (row II), and triple MTS mutant (rows III and IV) (scale bars: 10 μm).
(F) Fractionation of Flp-In TREx293 cell lysates overexpressing METTL8-F/H constructs into debris/nuclei, cytosol, and mitochondria. METTL8-F/H was detected by anti-HA, and TOMM20 served as mitochondrial marker.