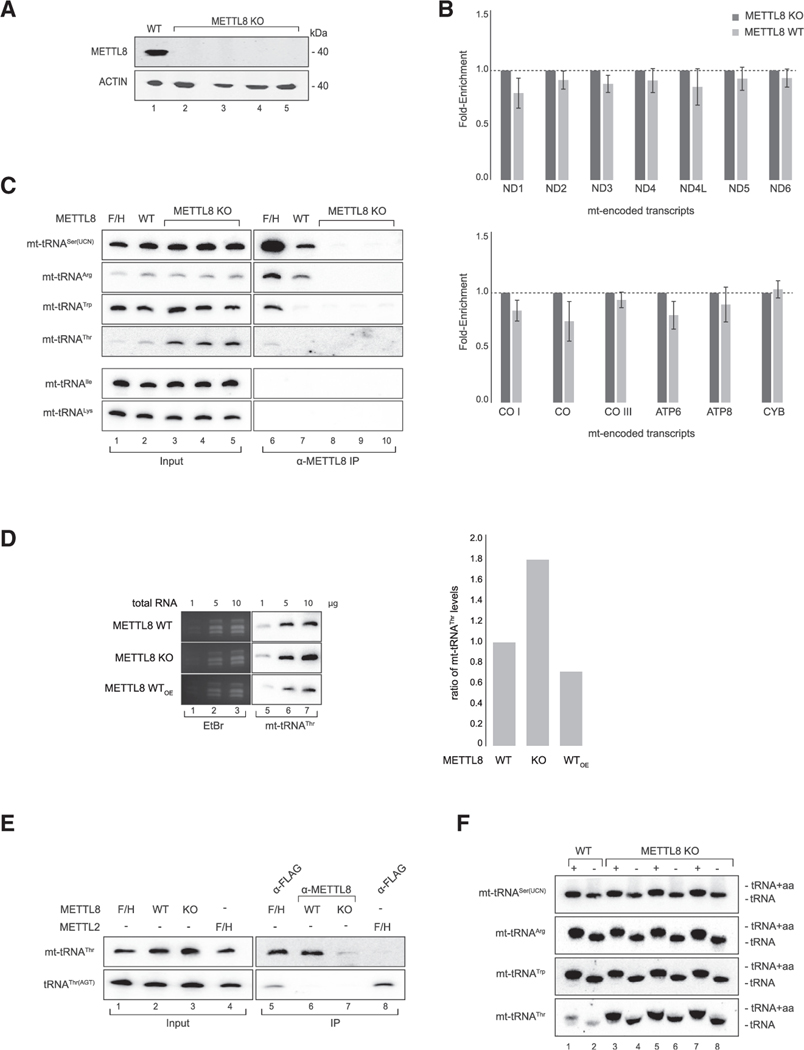
Figure 2. METTL8 interacts with mitochondrial tRNAs.
(A) Detection of METTL8 in Flp-In TREx293 METTL8 WT and METTL8 KO cell lines after anti-METTL8 IP. ACTIN served as the control.
(B) METTL8 RNA immunoprecipitation (RIP)-qPCR of METTL8 WT, and METTL8 KO Flp-In TREx293 cell lines. METTL8 KO cell line was used as control. Means ± standard error of the difference (SED) of three independent measurements are plotted.
(C) Northern blot analysis of RNAs co-immunoprecipitated with METTL8. METTL8 KO cell lines and 0.5 μg total RNA for input were used as controls.
(D) Northern blot analysis of mt-tRNAThr levels in overexpressing METTL8 WT, METTL8 WT, and KO cells. EtBr staining served as the loading control. Right: corresponding phosphor imaging quantification is shown.
(E) Northern blot analysis of RNAs co-immunoprecipitated with METTL8 and METTL2. METTL8-F/H and F/H-METTL2 were immunoprecipitated using anti-FLAG. 0.5 μg total RNA was used as the input control.
(F) Mitochondrial tRNA aminoacylation (+) and deacylation (−) levels in METTL8 WT and METTL8 KO cell lines.