This study demonstrates that ATM is essential for safe retroviral transduction in T-cells, and its lack poses a risk of chromosomal aberrations and potential leukemogenicity.
Abstract
Somatic variants in DNA damage response genes such as ATM are widespread in hematologic malignancies. ATM protein is essential for double-strand DNA break repair. Germline ATM deficiencies underlie ataxia–telangiectasia (A–T), a disease manifested by radiosensitivity, immunodeficiency, and predisposition to lymphoid malignancies. Patients with A–T diagnosed with malignancies have poor tolerance to chemotherapy or radiation. In this study, we investigated chimeric antigen receptor (CAR) T cells using primary T cells from patients with A–T (ATM−/−), heterozygote donors (ATM+/−), and healthy donors. ATM−/− T cells proliferate and can be successfully transduced with CARs, though functional impairment of ATM−/− CAR T-cells was observed. Retroviral transduction of the CAR in ATM−/− T cells resulted in high rates of chromosomal lesions at CAR insertion sites, as confirmed by next-generation long-read sequencing. This work suggests that ATM is essential to preserve genome integrity of CAR T-cells during retroviral manufacturing, and its lack poses a risk of chromosomal translocations and potential leukemogenicity.
Significance: CAR T-cells are clinically approved genetically modified cells, but the control of genome integrity remains largely uncharacterized. This study demonstrates that ATM deficiency marginally impairs CAR T-cell function and results in high rates of chromosomal aberrations after retroviral transduction, which may be of concern in patients with DNA repair deficiencies.
Introduction
Ataxia–telangiectasia (A–T) is a rare genetic disorder that is caused by pathogenic variants in the ATM gene, which is required for double-strand DNA (dsDNA) break repair. Patients are diagnosed during early childhood and present with cerebellar dysfunction, eye telangiectasia, and recurrent sinopulmonary infections and have a high incidence of lymphoid malignancies, both B cell– and T cell–derived (1–3). Patients with A–T have a short life expectancy, and the diagnosis of malignancy is associated with an even shorter overall survival (3). Current standard chemotherapy-based protocols are highly toxic to these patients, and no specific interventional guidelines have been set for patients with A–T diagnosed with lymphoma or leukemia (4).
T cell–engaging immunotherapy, which includes bispecific antibodies or chimeric antigen receptor (CAR) T cells, has led to high remission rates and is approved for B cell–derived malignancies (5–9). We sought to explore the potential feasibility of CAR T-cell therapy for A–T–derived malignancies, as such products have shown durable remissions in first and second relapse of B-cell lymphoma, commonly seen in this population and their application may reduce the need for cytotoxic chemotherapy. Immune deficiency is a well-described phenomenon in A–T. This is related to aberrant dsDNA repair during VDJ recombination in B-cell and T-cell development, leading to a paucity of B cells, T cells, naïve CD4+ cells, and immunoglobulins (3, 10, 11). CAR T-cell production also requires activation of autologous T cells and transduction with a retroviral or lentiviral vector encoding the CAR. Hence, there may be a risk following insertion of viral sequences into the genome of ATM−/− T cells.
Here, we studied the feasibility, efficacy, and toxicity of CAR T-cell production in cells collected from patients with A–T, representing a variety of pathogenic variants, most with a null protein function. We used matched heterozygous controls that carry the same ATM mutation in a single allele, as well as healthy controls. Altogether, this study provides data on retroviral transduction of CARs in ATM−/− and ATM+/− primary T cells, providing insights for modeling therapeutic viral integration in patients with DNA repair dysfunction.
Results
Feasibility of CAR transduction in ATM-deficient T cells
To assess the feasibility of CAR T-cell production in immune-deficient patients with A–T, we collected peripheral blood mononuclear cells (PBMC) from eight pairs of patients with A–T (ATM−/−) and their family members who served as heterozygous controls (ATM+/−). Patient details are presented in Supplementary Table S1. None of the patients had a prior cancer diagnosis or had received cytotoxic therapy. Cells from eight patients with A–T, eight heterozygous controls, and seven healthy controls were activated and transduced with a CD19 CAR. OKT3-activated untransduced cells of each genotype served as additional controls. This procedure resulted in the expansion of cells from all donors. The median fold expansion of ATM−/− CAR T cells was 2.9 times throughout the 10-day process compared with 7.8 times for ATM+/− and 10.7 times for ATM+/+ CAR T cells (Fig. 1A and B). The median CD3+ content in the starting material was 24% in ATM−/− and 43% in ATM+/− PBMCs. By the end of the expansion, the CD3+ percentage increased to 95.8 and 97.9, respectively (Fig. 1C), confirming a high T-cell purity in all products. The median CAR transduction efficacies were 63%, 75%, and 79% in ATM−/−, ATM+/−, and ATM+/+ products, respectively (Fig. 1D and E). CD4 and CD8 subpopulations were similar across products (Fig. 1F). Of note, viability of the ATM−/− CAR T cells was lower than that of the controls at the end of the production (Supplementary Fig. S1A and S1B).
Figure 1.
CAR T-cell production in patients with A–T. PBMCs from patients (ATM−/−), heterozygous controls (ATM+/−), and healthy donors (ATM+/+) were activated with OKT3 and IL2, transduced with a CD19 CAR T cell, and expanded in culture for 10 days. A, Fold change representing cell growth of ATM−/− (red) and ATM+/− (blue) cells during production. Median and IQR are shown. *, P < 0.05; **, P < 0.01. B, Summary of day 10 fold change of ATM−/−, ATM+/−, and ATM+/+ products. Line depicts the median in each group. C, CD3+ content in peripheral blood and CAR T products of ATM−/− (red) and ATM+/− (blue). Each blood sample/product is represented by a dot. Lines represent matched blood-product samples from patients and controls. D, Flow cytometry histograms representative of CAR transduction in ATM−/− (red), ATM+/− (blue), and ATM+/+ (gray) products and untransduced (UT) control T cells. E, Summary of CAR transduction efficacy. Each product is represented by a dot. Line depicts the median in each group. F, Average CD4+ and CD8+ subsets in CAR T final products. Error bars represent SD. N = 4 per group. In A–E, N = 8 samples for ATM−/−, n = 8 for ATM+/− products, and n = 7 for ATM+/+ products. A Mann–Whitney nonparametric test was used to compare each of the two groups.
ATM-deficient CAR T cells are functionally impaired compared with heterozygotes or normal controls
To test the in vitro efficacy of ATM−/− and ATM+/− CAR T cells, a coculture with Nalm6 CD19+ cells was performed. During a short cytotoxicity assay, CD19 CAR T cells showed a rapid induction of annexin expression in all targets after 2.5 hours and subsequent target killing compared with untransduced controls. However, the specific cytotoxicity of ATM−/− CAR T cells was lower than that of ATM+/− controls and ATM+/+ controls (Fig. 2A). We observed a lower viability of CAR T products from ATM−/− donors when resting (Supplementary Fig. S1A and S1B) and after activation (Supplementary Fig. S1C). We analyzed annexin V expression on CAR transduced and untransduced T cells after activation with Nalm6 cells. ATM−/− CAR T-cells showed increased staining for annexin V in some experiments (Supplementary Fig. S2A and S2B) compared with ATM+/− and ATM+/+ T cells, especially when comparing paired ATM+/− and ATM−/− CAR T-cells from family members carrying a heterozygous or homozygous mutation (Supplementary Fig. S2C). This suggests the susceptibility of ATM−/− CAR T cells to higher activation-induced cell death, but it was not observed in all donors.
Figure 2.
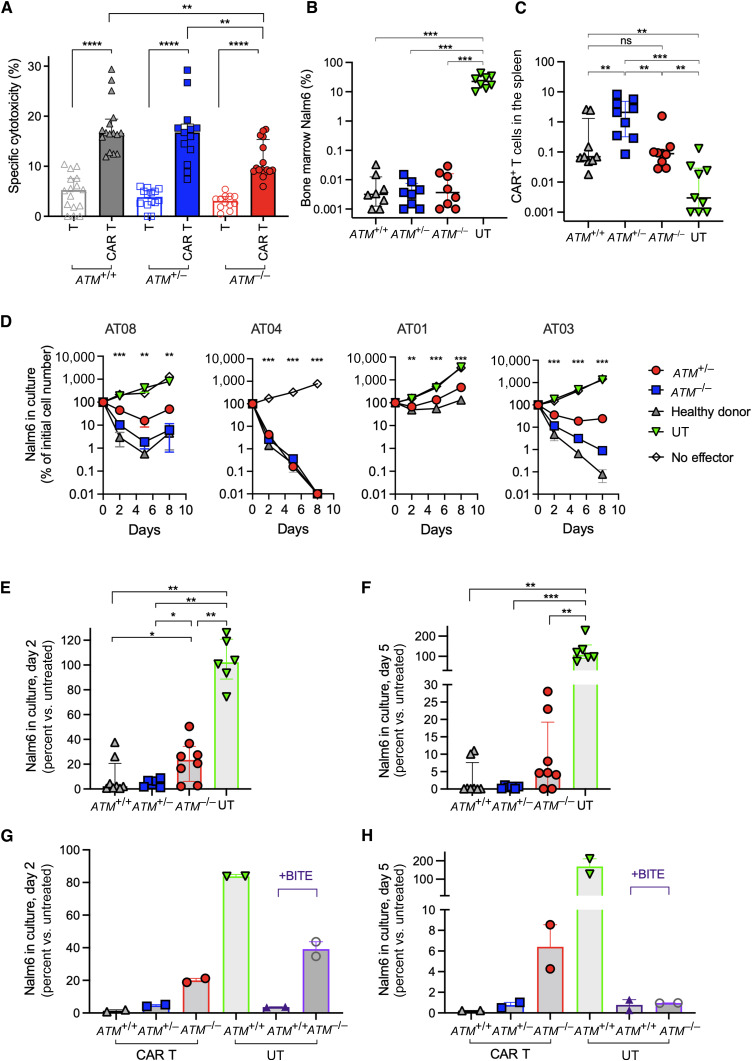
In vitro and in vivo activity of CD19 CAR T cells. A, CAR T cells (colored) or untransduced T cells (clear) from healthy donor products (ATM+/+, gray), heterozygous ATM+/− products (blue), or ATM−/− products (red) were cocultured with CellTrace Violet –stained Nalm6 cells. The annexin V level was assessed gated on CellTrace Violet–positive cells after 2.5 hours. Each dot represents an independent experimental result. Products from all donors were used at least once (n = 8 ATM−/−, n = 8 ATM+/−, and n = 7 ATM+/+ donors). B and C, For in vivo studies, Nalm6-inoculated NSG mice were treated with 2 × 106 CD19 CAR T cells from different donors. B, Bone marrow leukemic cells at 14 days post–CAR T-cell administration, gated on murine CD45− and human CD10+ cells. C, Spleen CAR T-cell percentage, gated on murine CD45− and human CD3+ CAR+, out of all mononuclear cells. D–H, Stress experiment of CD19 CAR T cells from different donors against Nalm6 in an E:T ratio of 1:16 for 8 days. Viable Nalm6 cells (gated on viable CD10+ cells) in culture are assessed at days 2, 5, and 8 by flow cytometry. D, Number of viable Nalm6 cells in culture of different pairs of ATM−/− cells (red circles), ATM+/− cells (from each patient’s heterozygous control, blue squares), healthy donor–derived cells (ATM+/+, gray triangles), and untransduced (UT, green triangles) or no effector (clear) controls. Cell number is shown as a percentage of the initial Nalm6 cell number in the coculture. One-way ANOVA was used to calculate P values per time point. E and F, Summary of Nalm6 percent compared with untreated cells on days 2 (E) and 5 (F) of coculture. Each dot represents an independent experiment result. G and H, Similar coculture was performed with CAR T cells from healthy donors (ATM+/+, gray triangles), ATM+/− heterozygous controls (blue squared), and ATM−/− (red circles); and untransduced cells from healthy donor (ATM+/+, green), untransduced ATM+/+ T cells with blinatumomab (BITE, purple triangles), and ATM−/− untransduced cells with blinatumomab (BITE, purple circles). Blinatumomab was added on days 0, 2, and 5 of coculture at 50 ng/mL/well. Each dot represents an independent experiment result from a different donor (n = 2). Live target cells (CD10+) were counted on days 2 (G) and 5 (H) and measured as a percentage of the initial number of Nalm6 cells cultured, with a low number representing enhanced clearance by T cells. BITE= Bispecific T-cell engager. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
We next assessed the in vivo efficacy of ATM−/− CAR T cells in NSG mice inoculated with Nalm6 cells. Four days after leukemia injection, mice were treated with ATM−/−, ATM+/−, or ATM+/+ CAR T cells. After 14 days, leukemic cells were cleared in all mice regardless of the donor source (Fig. 2B). The analysis of in vivo proliferation of CAR T-cells in the spleens of treated mice showed that CAR T-cell proliferation was similar in ATM−/− and ATM+/+ donor CAR T cells and higher in ATM+/− CAR T cells (Fig. 2C).
Previous experiments showed marginal inferiority of ATM−/− CAR T cells compared with controls. Although patient-derived material was limited, we further conducted an in vitro stress experiment in which CD19 CAR T-cells from different donors were cocultured with Nalm6 cells at an effector-to-target (E:T) ratio of 1:16. In four independent experiments, most donors failed to completely clear leukemic cells after 8 days. Using four different donor trios (ATM−/−, ATM+/−, and ATM+/+ donors), we observed that in three of the four experiments, ATM−/− CAR T cells were inferior to the ATM+/+ or ATM+/− controls in achieving leukemic cell clearance (Fig. 2D). Altogether, ATM−/− CAR T cells were less efficient in clearing leukemic cells in cocultures compared with controls on day 2 (Fig. 2E) or day 5 (Fig. 2F) of coculture.
Bispecific engagers have similar efficacy to CAR T cells in ATM-deficient products
Furthermore, we also tested whether the bispecific T-cell engager blinatumomab, which also targets CD19, may lead to leukemic cell clearance in these patients. In a similar assay, the addition of blinatumomab to untransduced T cells from healthy ATM+/+ donors led to clearance of leukemia. This was comparable with leukemic cell clearance by CAR T-cells from healthy donors or ATM+/− CAR T-cells. On day 2 of coculture, ATM−/− CAR T cells as well as untransduced ATM−/− T cells with blinatumomab failed to completely deplete leukemic cells in the culture (Fig. 2G). However, after 5 days, the elimination of leukemic cells by the addition of blinatumomab to untransduced T cells was similar in ATM−/− and ATM+/+ donor T cells, and the elimination effect in both cells was similar to that in healthy donor or ATM+/− CAR T cells (Fig. 2H).
Retroviral integration in ATM-deficient cells leads to chromosomal aberrations
Retroviral vectors encoding the CARs integrate into the T-cell genome. The retroviral enzymes require dsDNA repair machinery for postintegration repair (12). We successfully transduced ATM−/− T cells with a retroviral-based CAR (Fig. 1D and E) as well as GFP (Supplementary Fig. S3). We next applied cytogenetic analysis on ATM−/−, ATM+/−, and healthy donor CAR T products and untransduced activated T cells. Altogether, 385 cells were analyzed from 37 products. All ATM−/− CAR T-cell products had at least one cytogenetic aberration, and clonal aberrations were seen in four of seven products (Fig. 3A–D; Supplementary Table S2). The most common cytogenetic abnormalities involved chromosomes 7 and 14. The incidence of chromosomal aberrations in ATM−/− CAR T cells was significantly higher than their incidence in ATM+/− or ATM+/+ products (Fig. 3B and C). Overall, 27/48 cells analyzed from seven ATM−/− CAR T products had cytogenetic aberrations, compared with 5/82 cells from ATM−/−untransduced products, 3/75 from ATM+/− CAR T cells, and 0/47 from ATM+/+ healthy donor–derived CAR T-cell products (one-way ANOVA, P < 0.001). We next transduced ATM−/− T cells with a GFP-encoding or CD19 CAR, both on an MSGV backbone, to assess genomic instability following non-CAR retroviral transduction. Indeed, ATM−/− cells had chromosomal aberrations following either GFP or CD19 CAR transduction (Supplementary Table S3).
Figure 3.
Cytogenetic analysis of CAR T and control products. A, Representative images of karyotypes from a cell taken from (i) a healthy donor ATM+/+ CAR T product, (ii) an ATM+/− CAR T product, (iii) an untransduced activated ATM−/− T cells, and (iv) an ATM−/− CAR T-cell product. Chromosomes are G-banded and numbered. M, marker chromosome. B–D, Column summary of 385 cells analyzed from 37 products: (B) Each dot represents a T-cell product analyzed for cytogenetic events. The number of abnormal karyotypes per product is shown on the Y axis. Six healthy donor ATM+/+ untransduced activated T cells (T) or CAR T-cell (CAR T) products were analyzed. One of six untransduced products had nine cytogenetic aberrations, and all the CAR products had no aberrations. In ATM+/−products, two of the untransduced cell products had an aberrant karyotype, as well as one product that was CAR-transduced. In ATM−/− products, four of seven untransduced products had cytogenetic aberrations, and all ATM−/− CAR T products had aberrations. P value was calculated using one-way ANOVA. C, Plot representing individual cells analyzed in all products as in B. D, Plot per product as in B, representing clonal events, in which an aberration was shared in more than one cell analyzed per product. *, P < 0.05; ***, P < 0.001.
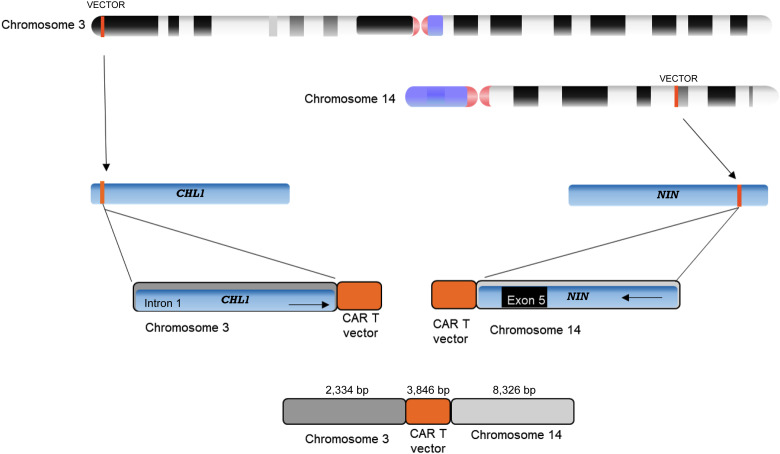
The increased incidence of cytogenetic events in ATM−/− CAR T cells suggested that the retroviral integration itself is responsible for chromosomal translocations in these cells, rather than the T-cell activation process. To substantiate this assumption, we sequenced genomic DNA from two products (from the donors AT06 and AT08) using the Pacific Biosciences single-molecule long-read analysis platform. Circular consensus sequencing reads were aligned to the human genome (hg38) together with the vector sequence using the pbmm2 analysis tool. In order to detect insertion and translocation events, we filtered for reads that were aligned to the vector as well to a genomic sequence. We could find three main types of these reads (see illustration in Supplementary Fig. S4): (i) reads with partial mapping to the vector at the start or end of the read and to the human genome on the rest of the read. Using these reads, we cannot conclude if the genomic event is insertion of the vector in a specific position or an insertion resulting in a translocation. (ii) Reads with full mapping to the vector sequence in the middle of the read together with reads mapping to a specific position in the genome. In these cases, junctions of the viral sequences with genomic DNA were identified in both ends of the construct. These reads suggest an insertion event of the vector at this genomic position. (iii) Reads with full mapping to the vector in the middle of the read, whereas the two ends of the read are mapped to two different genomic locations. These reads suggest translocation events related to the vector insertion. A full list of the reads found to be aligned to the vector is detailed in Supplementary Table S4. Of these, two reads supporting vector-related translocation (type 3) were found in the data (Supplementary Table S4; Supplementary Fig. S5). The first one has 2,334 bp aligned to chromosome 3, followed by 3,846 bp alignment to the vector, and finally 8,325 bp aligned to chromosome 14 (Fig. 4). The second read has 1,861 bp aligned to chromosome 7, followed by 3,856 bp aligned to the vector. Interestingly, in this read, we found an additional 1,265-bp sequence aligned to the vector, which is probably a duplication, and finally 4,965 bp aligned to chromosome 20.
Figure 4.
Illustration of a translocation event resulting in a read of 14.5 kb from an ATM−/− CAR T product. The upper 2,334 bases align to the chromosome 3 sequence (includes part of CHL1 intron 1), followed by a 3,846-bp sequence that aligns to the CAR T vector, and the 8,326 bp downstream part of the read aligns to the chromosome 14 sequence (includes exon 5 and part introns 6 and 4 of the NIN gene).
Discussion
The data in this study provide insights into genotoxicity in retrovirally transduced T cells harboring defects in dsDNA repair mechanisms. These insights are substantiated through the analysis of multiple patient samples, each featuring distinct pathologic variants of the ATM gene (the majority of which result in complete loss of protein function), as well as data from paired heterozygous controls (representing varying degrees of dsDNA repair deficiency) and healthy donors, thus providing comprehensive validation of our findings.
Our initial goal was to assess the feasibility of cellular immunotherapy in patients with A–T who have a high incidence of lymphoid malignancies and are intolerant to chemotherapy. Despite known immune deficiency in patients with A–T, successful production of CAR T cells from all patients and controls was achieved, though with a slightly lower expansion and lower transduction efficacy and a lower viability in ATM−/− CAR T cells. A functional disadvantage was seen in vitro under stress conditions, with marginally increased activation-induced cell death in ATM−/− cells, yet still successful targeting of CD19-expressing target cells was achieved.
Overall, despite a known lower infectivity of retroviruses in ATM−/− cells (12), transduction efficacy was still achieved in a significant percentage of cells, and activity against the CAR targets, both in vivo and in vitro, was maintained. Only in a stress model, using an E:T ratio of 1:16, the ATM−/− CAR T cells showed inferior ability to controls. Although the activation was CAR-mediated and not T-cell receptor (TCR)–mediated, the TCR diversity was irrelevant to this effect. There is a known lack of CD4+ and naïve cells in patients with A–T, and these subpopulations were found to be significant for CAR T activation and long-term maintenance (13). There was a lower CD4 percentage in all CAR T products regardless of the donor. Our findings resulted from relatively short assays, thus not informative about long-term effects and clonal selection occurring in patients. Due to limited patient-derived material, we could not conduct long-term in vivo experiments that require a higher CAR T-cell dose use to assess long-term functionality. Nevertheless, functional differences were repeatedly observed in cell viability and under stress conditions.
We investigated alternative targeting methods using bispecific antibodies. Blinatumomab, a bispecific CD19–CD3 engager sharing target with the CAR used in this study, was evaluated in the presence of untransduced T cells, and compared with CD19 CAR T-cells for efficacy. Following 5-day coculture, bispecific antibodies resulted in similar target elimination when applied on ATM−/− or healthy donor ATM+/+ T cells. This was comparable with the target elimination achieved with CAR T-cells from healthy or ATM+/− donors. The kinetics of leukemic cell clearance with the addition of blinatumomab were lower than those with CAR T cells in both ATM−/− and ATM+/+ cells, as seen by incomplete leukemic cell clearance on day 2. The inherent CD28 costimulation of the CAR may provide faster killing kinetics than mere CD3 engagement. Bispecific antibodies activate T cells but do not require viral integration. Thus, they do not pose more risk of genotoxicity than activated T cells. The rate of chromosomal aberrations per cell was significantly lower in ATM−/− untransduced cells compared with ATM−/− CAR T-cells and was similar to ATM+/− and ATM+/+ controls. Promising success of bispecific antibodies targeting CD19 or CD20 (14), for example, may lead to their utilization in patients with A–T who are prone to severe chemotherapy-associated adverse events.
ATM −/− lymphocytes are prone to develop chromosomal aberrations, frequently involving illegitimate recombination of TCR genes. A main finding in our work was the frequent cytogenetic abnormalities observed in ATM−/− CAR T-cell products, not necessarily involving the TCR genes, further raising a concern about the use of genetically modified cells in patients with A–T. Early trials showed that retroviral transduction of hematopoietic stem cells was associated with leukemogenicity in patients with X-linked severe combined immune deficiency and Wiskott–Aldrich syndrome (15, 16), as a result of viral integration in leukemogenic regions such as LMO2. Long-term follow-up of patients undergoing T-cell retroviral transduction showed no risk of genetic integration or leukemogenicity (17, 18). Recently, concerns over T-cell malignancies in CAR T–treated patients have emerged (19, 20), in contrary to the long safety data previously published. The cytogenetic analysis performed in this study showed alterations in some CAR T-cell and activated T-cell products, but alarmingly, all ATM−/− CAR T products had cytogenetic abnormalities, with many of these being clonal abberations. This occurred during a relatively short culture time, without the longer time period needed for clonal selection associated with full leukemic transformation. However, the high rate of alterations and the common involvement of chromosomes 7 and 14 are of great concern, as these are commonly involved in translocations in T-cell malignancies occurring in patients with A–T (1). The genomic alterations were not a result of excessive T-cell activation because ATM−/− untransduced activated controls did not have such a high rate of chromosomal aberrations. CAR T-cell products from ATM+/− heterozygotes related to the patients with A–T, representing intermediate ATM dysfunction, did not have a high rate of chromosomal translocations, suggesting that the normal product from the functioning allele is sufficient to prevent enhanced accumulation of aberrations. Retroviral transduction with GFP also led to chromosomal aberrations in ATM−/− T cells, though assessed on a small number of cells. We conclude that these events are highly abundant following the combination of retroviral transduction and complete lack of ATM function. The use of multiple patient samples, each harboring a different ATM variant (most with a null protein function), and the data from paired heterozygous controls strengthen our conclusion. Moreover, long-read next-generation sequencing identified two events in which the CAR vector resided within a chromosomal translocation junction. This work further confirms a significant role of ATM in retroviral integration (21). The increase in cell death after retroviral transduction, reported previously (21) and observed in some of our samples, raises the concern whether the CAR provides additional survival signaling to prevent the cell from early apoptosis, thus increasing the potential leukemogenic risk.
Patients with defective DNA repair mechanisms, A–T as an example, are at high risk for malignancies. A high rate of germline ATM variants was reported in patients with chronic lymphocytic leukemia (22), mantle cell lymphoma, and other B-cell malignancies, further expanding the relevance of our findings to a larger patient population. Therapeutic virally transduced T cells are increasingly used, and it may be worthwhile to screen for germline mutations associated with impairment in DNA repair. If confirmed, alternatives to genetically manipulated cells, such as bispecific antibodies, may have a similar efficacy with an improved safety profile.
Methods
Patient samples and ethical approval
All patient samples were collected according to an institutional review board–approved protocol (SMC-19-6025). A signed written informed consent was obtained from all patients. Samples were taken from the peripheral blood of patients during a routine visit at the national A–T clinic Sheba Medical Center. Approximately 15 mL of blood was drawn from ATM−/− patients and their first-degree relatives known to be carriers of the mutation on a single allele (ATM+/−, usually one of the parents). All animal experiments were approved by the Institutional Ethical Review Process Committee and were performed under Israel Institutional Animal Care and Use Committee approval (no. 1216/19).
Cell culture
The Nalm6 acute lymphoblastic leukemia cell line was kindly provided by Steve Rosenberg of the NCI and grown under standard culture conditions, as previously reported (23, 24). The cell line was tested negative for Mycoplasma using the Hy-Mycoplasma PCR assay. A T-cell medium for activation and transduction included RPMI 1640 supplemented with 10% FBS, 2 mmol/L L-Glu, 100 U/mL penicillin, 100 μg/mL Streptomycin (all from Sarorius, Beit Haemek), and IL2 (100 IU/mL, Clinigen, Staffordshire). 293T cells used for viral production were cultivated in D10 media consisting of DMEM high glucose supplemented with 10% FBS, 2 mmol/L L-glutamate, sodium pyruvate, HEPES buffer 0.1 mol/L, and nonessential amino acid solution (all from Sartorius, Beit Haemek).
CAR production
PBMCs were isolated using centrifugation on Lymphoprep density gradients (Fresenius Kabi Norge, Oslo) and were activated in a T-cell medium with 100 IU/mL IL2 and anti-CD3 Ab OKT3 50 ng/mL (Invitrogen). On day 2 of culture, activated cells were transduced with CD19 CAR, containing an FMC63-CD28-CD3ζ vector, on an MSGV retroviral backbone, as previously reported (23) and used in our clinical trial (24). For this purpose, nontissue culture–treated six-well plates (Falcon) were precoated with 20 µg RetroNectin per well (T100B, Takara) at 4°C overnight followed by a 20-minute incubation with 2.5% BSA (Caisson Labs, BSA fraction V). The plates were loaded with 5 mL viral supernatant per well and centrifuged at 2,000 g for 2 hours at 32°C. Following centrifugation, the supernatant was collected leaving only 1 mL/well. Then, the plates were seeded with 2 × 106 OKT3-activated PBMCs/well, centrifuged at 1,000 g for 20 minutes at 32°C, and incubated overnight at 37°C. The following day, the cells were collected from transduction plates and further expanded in IL2-containing T-cell medium until day 10. For GFP transduction, we used EGFP plasmid cloned into the MSGV1 retroviral vector (provided by Gal Cafri and Yardena Samules from the Weizmann Institute of Science). T cells were transduced using the same retroviral transduction protocol as CAR.
Short cytotoxicity assay
Nalm6 cells were stained with CellTrace Violet according to the manufacturer’s instructions and seeded at 180,000 cells/well in a 96-well plate. CAR T cells were then added at 20,000 cells/well all diluted to 50% transduced cells using T cells from the untransduced control T-cell culture (to a total of 40,000 T cells/well) to provide a final E:T ratio of 1:10. The total culture volume was 200 µL/well. After 2.5 hours at 37°C, the cells were collected from wells and stained for apoptosis using Annexin V-Cy5 reagent (Biovision). CellTrace-positive target cells were assessed for annexin V staining by flow cytometry.
Stress long-term cytotoxicity
Nalm6 target cells were seeded at 250,000 cells/well in a 24-well plate. A total of 15,625 CAR+ cells were added to maintain a desired E:T ratio of 1:16. Untransduced T cells were added to achieve a total of 100,000 T cells per well to control for different transduction efficacy percentages in products from different donors. The total culture volume was 1 mL/well, and 100 µL was sampled from the culture, counted, and assessed for the presence of CD10+ and CD3+ cells by flow cytometry. When indicated, blinatumomab (Amgen) was added at 50 ng/mL to each well on days 0, 2, and 5 of coculture.
In vivo models
For all in vivo experiments, 8- to 15-week-old NOD/SCID IL2Rγ− (NSG) female mice (The Jackson Laboratory) were used. Mice were injected with 0.4 × 106 Nalm6 cells via tail vein. Four days later, effector cells were injected intravenously at 2.0 × 106 CAR T cells/mouse. The mice were monitored and sacrificed after 14 days. Their spleen and bone marrow were harvested for further examination.
Cytogenetic analysis
Karyotype analysis of all the cultures was performed using a conventional G-banding technique. Briefly, 2 × 106 cells from transduced and untransduced cultures were activated overnight with phytohemagglutinin. For metaphase preparation, colcemid (0.2 μg/mL, Biological Industries, Israel) was added for 0.5 hours, followed by treatment with a hypotonic solution (0.075 mol/L KCl, Biological Industries, Israel) at 37°C for 10 minutes and fixation in methanol/acetic acid (3:1, Merck) at room temperature for 25 minutes. Karyotype analysis of trypsin–Giemsa–banded metaphase spreads was performed on the GenASIs Scan & Analysis platform (Applied Spectral Imaging, Carlsbad, USA). Karyotypes were classified according to the 2016 Committee on Human Cytogenetic Nomenclature (25).
PacBio sequencing
Genomic DNA was extracted from cell culture samples using the Nanobind CBB HMW DNA extraction kit (102-301-900), following the manufacturer’s protocol. The extracted DNA was quantified using a Qubit fluorometer and NanoDrop spectrophotometer and assessed for size using the Genomic DNA Tapestation system. For library preparation, genomic DNA was sheared into fragments using Covaris g-TUBE (520079), followed by DNA damage repair and end-repair. SMRTbell adapters were ligated onto the repaired DNA fragments according to the SMRTbell prep kit 3.0 (PacBio, 102-182-700). The quality of the resulting libraries was assessed using the Genomic DNA Tapestation system. SMRT sequencing was performed on the PacBio Sequel IIe platform using SMRT cells at 8 mol/L and Sequel II sequencing kit 2.0, according to the manufacturer’s recommendations. The movies were collected for 30 hours, generating HiFi long-read sequencing data.
Flow cytometry
Flow cytometry was performed on Beckman Coulter Gallios. For detection of the CD19 CAR, we used the biotinylated Miltenyi CD19 CAR detection reagent (Cat# 130-115-965) and streptavidin-APC as a secondary Ab (BioLegend, Cat# 405207). Antibodies anti–human CD3–FITC (Cat# 130-113-138), anti–human CD19-PE (Cat# 130-113-646), anti–human CD10-PE-Cy7 (Cat# 130-114-504), and anti–mouse CD45 APC-Cy7 (Cat# 130-110-800; all from Miltenyi) were used for additional staining and Ghost Red 780 viability dye (Tonbo Biosciences, 13-0865-T100) for dead cell exclusion. Analysis was carried out using FlowJo analysis software V10.
Statistical analysis
All statistical analyses were carried out on GraphPad Prism software. SDs were calculated from technical repeats, and SEM was plotted with biological repeats. Unless otherwise specified, group comparative analysis of continuous variables was carried out using a nonparametric test (Mann–Whitney) because of small numbers per group. Analysis of categorical events was performed using the χ2 test, and P value was calculated using the Fisher exact test.
Data availability
PacBio sequencing data generated for this study have been deposited in the European Nucleotide Archive accession PRJEB75002.
Supplementary Material
Patient characteristics
Detailed cytogenetic analysis of CAR-T and untransduced activated cells
Cytogenetic analysis of ATM-/- transduced with GFP or CAR
Alignment details for reads found to be aligned to the CAR vector
Viability of CAR-T cells from ATM+/+, ATM+/- and ATM-/- donors at the end of production
Activation-induced cell death in samples from ATM+/+, ATM+/- and ATM-/- donors.
Retroviral transduction of GFP or CAR in ATM+/+ and ATM-/- T-cells
Illustration demonstrating structure of different type of reads aligned to the CAR vector
Illustration demonstrating structure of two reads supporting vector-related translocation
Acknowledgments
The authors would like to thank Diana Bar for clinical assistance and Drs. Gal Cafri and Orit Itzhaki for assistance with methodologies and reagents. We are grateful to the patients and their families who participated in the study through the national A–T clinic. This work was supported by the Action for A-T grant (EJ) and Dotan Research Center in Hematologic Malignancies grant (G. Rechavi and E. Jacoby).
Footnotes
Note Supplementary data for this article are available at Blood Cancer Discovery Online (https://bloodcancerdiscov.aacrjournals.org/).
Authors’ Disclosures
E. Jacoby reports grants from Action for A-T and Dotan Research Center in Hematologic Malignancies during the conduct of the study. No disclosures were reported by the other authors.
Authors’ Contributions
M. Rozenbaum: Formal analysis, investigation, methodology. R. Fluss: Formal analysis, investigation, methodology, writing–review and editing. V. Marcu-Malina: Formal analysis, investigation, methodology, writing–review and editing. I. Sarouk: Conceptualization, investigation. A. Meir: Investigation. S. Elitzur: Conceptualization, writing–review and editing. T. Zinger: Formal analysis, investigation. J. Jacob-Hirsch: Formal Analysis, methodology, writing-review, editing. E. Glick Saar: Formal analysis, supervision, methodology. G. Rechavi: Conceptualization, resources, formal analysis, supervision, writing–review and editing. E. Jacoby: Conceptualization, resources, formal analysis, supervision, funding acquisition, investigation, methodology, writing–original draft, writing–review and editing.
References
- 1. Taylor A, Metcalfe J, Thick J, Mak Y. Leukemia and lymphoma in ataxia telangiectasia. Blood 1996;87:423–38. [PubMed] [Google Scholar]
- 2. Bielorai B, Fisher T, Waldman D, Lerenthal Y, Nissenkorn A, Tohami T, et al. Acute lymphoblastic leukemia in early childhood as the presenting sign of ataxia-telangiectasia variant. Pediatr Hematol Oncol 2013;30:574–82. [DOI] [PubMed] [Google Scholar]
- 3. van Os NJH, Jansen AFM, van Deuren M, Haraldsson A, van Driel NTM, Etzioni A, et al. Ataxia-telangiectasia: immunodeficiency and survival. Clin Immunol 2017;178:45–55. [DOI] [PubMed] [Google Scholar]
- 4. Elitzur S, Shiloh R, Loeffen J, Pastorczak A, Takagi M, Bomken S, et al. ATM germline pathogenic variants affect treatment outcomes in children with acute lymphoblastic leukemia/lymphoma and ataxia telangiectasia. Blood 2023;142(Suppl 1):520. [DOI] [PubMed] [Google Scholar]
- 5. Maude SL, Laetsch TW, Buechner J, Rives S, Boyer M, Bittencourt H, et al. Tisagenlecleucel in children and young adults with B-cell lymphoblastic leukemia. N Engl J Med 2018;378:439–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Neelapu SS, Locke FL, Bartlett NL, Lekakis LJ, Miklos DB, Jacobson CA, et al. Axicabtagene ciloleucel CAR T-cell therapy in refractory large B-cell lymphoma. N Engl J Med 2017;377:2531–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Abramson JS, Palomba ML, Gordon LI, Lunning MA, Wang M, Arnason J, et al. Lisocabtagene maraleucel for patients with relapsed or refractory large B-cell lymphomas (TRANSCEND NHL 001): a multicentre seamless design study. Lancet 2020;396:839–52. [DOI] [PubMed] [Google Scholar]
- 8. Schuster SJ, Bishop MR, Tam CS, Waller EK, Borchmann P, McGuirk JP, et al. Tisagenlecleucel in adult relapsed or refractory diffuse large B-cell lymphoma. N Engl J Med 2019;380:45–56. [DOI] [PubMed] [Google Scholar]
- 9. Neelapu SS, Dickinson M, Munoz J, Ulrickson ML, Thieblemont C, Oluwole OO, et al. Axicabtagene ciloleucel as first-line therapy in high-risk large B-cell lymphoma: the phase 2 ZUMA-12 trial. Nat Med 2022;28:735–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bredemeyer AL, Sharma GG, Huang CY, Helmink BA, Walker LM, Khor KC, et al. ATM stabilizes DNA double-strand-break complexes during V(D)J recombination. Nature 2006;442:466–70. [DOI] [PubMed] [Google Scholar]
- 11. Driessen GJ, Ijspeert H, Weemaes CMR, Haraldsson Á, Trip M, Warris A, et al. Antibody deficiency in patients with ataxia telangiectasia is caused by disturbed B- and T-cell homeostasis and reduced immune repertoire diversity. J Allergy Clin Immunol 2013;131:1367–75.e9. [DOI] [PubMed] [Google Scholar]
- 12. Sakurai Y, Komatsu K, Agematsu K, Matsuoka M. DNA double strand break repair enzymes function at multiple steps in retroviral infection. Retrovirology 2009;6:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Melenhorst JJ, Chen GM, Wang M, Porter DL, Chen C, Collins MA, et al. Decade-long leukaemia remissions with persistence of CD4+ CAR T cells. Nature 2022;602:503–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Falchi L, Vardhana SA, Salles GA. Bispecific antibodies for the treatment of B-cell lymphoma: promises, unknowns, and opportunities. Blood 2023;141:467–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hacein-Bey-Abina S, Hauer J, Lim A, Picard C, Wang GP, Berry CC, et al. Efficacy of gene therapy for X-linked severe combined immunodeficiency. N Engl J Med 2010;363:355–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Braun CJ, Boztug K, Paruzynski A, Witzel M, Schwarzer A, Rothe M, et al. Gene therapy for Wiskott–Aldrich syndrome-long-term efficacy and genotoxicity. Sci Transl Med 2014;6:227ra33. [DOI] [PubMed] [Google Scholar]
- 17. Scholler J, Brady TL, Binder-Scholl G, Hwang W-TT, Plesa G, Hege KM, et al. Decade-long safety and function of retroviral-modified chimeric antigen receptor T cells. Sci Transl Med 2012;4:132ra53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hsieh EM, Myers RM, Yates B, Annesley C, John S, Taraseviciute A, et al. Low rate of subsequent malignant neoplasms after CD19 CAR T-cell therapy. Blood Adv 2022;6:5222–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Verdun N, Marks P. Secondary cancers after chimeric antigen receptor T-cell therapy. N Engl J Med 2024;390:584–6. [DOI] [PubMed] [Google Scholar]
- 20. Levine BL, Pasquini MC, Connolly JE, Porter DL, Gustafson MP, Boelens JJ, et al. Unanswered questions following reports of secondary malignancies after CAR-T cell therapy. Nat Med 2024;30:338–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lau A, Swinbank KM, Ahmed PS, Taylor DL, Jackson SP, Smith GCM, et al. Suppression of HIV-1 infection by a small molecule inhibitor of the ATM kinase. Nat Cell Biol 2005;7:493–500. [DOI] [PubMed] [Google Scholar]
- 22. Lampson BL, Gupta A, Tyekucheva S, Mashima K, Petrácčková A, Wang Z, et al. Rare germline ATM variants influence the development of chronic lymphocytic leukemia. J Clin Oncol 2023;41:1116–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rozenbaum M, Meir A, Aharony Y, Itzhaki O, Schachter J, Bank I, et al. Gamma-delta CAR-T cells show CAR-directed and independent activity against leukemia. Front Immunol 2020;11:1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Itzhaki O, Jacoby E, Nissani A, Levi M, Nagler A, Kubi A, et al. Head-to-head comparison of in-house produced CD19 CAR-T cell in ALL and NHL patients. J Immunother Cancer 2020;8:e000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McGowan-Jordan J, Simons A, Schmid M, editors. ISCN 2016: An international system for human cytogenomic nomenclature. Basel (Switzerland): S. Karger AG; 2016. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Patient characteristics
Detailed cytogenetic analysis of CAR-T and untransduced activated cells
Cytogenetic analysis of ATM-/- transduced with GFP or CAR
Alignment details for reads found to be aligned to the CAR vector
Viability of CAR-T cells from ATM+/+, ATM+/- and ATM-/- donors at the end of production
Activation-induced cell death in samples from ATM+/+, ATM+/- and ATM-/- donors.
Retroviral transduction of GFP or CAR in ATM+/+ and ATM-/- T-cells
Illustration demonstrating structure of different type of reads aligned to the CAR vector
Illustration demonstrating structure of two reads supporting vector-related translocation
Data Availability Statement
PacBio sequencing data generated for this study have been deposited in the European Nucleotide Archive accession PRJEB75002.