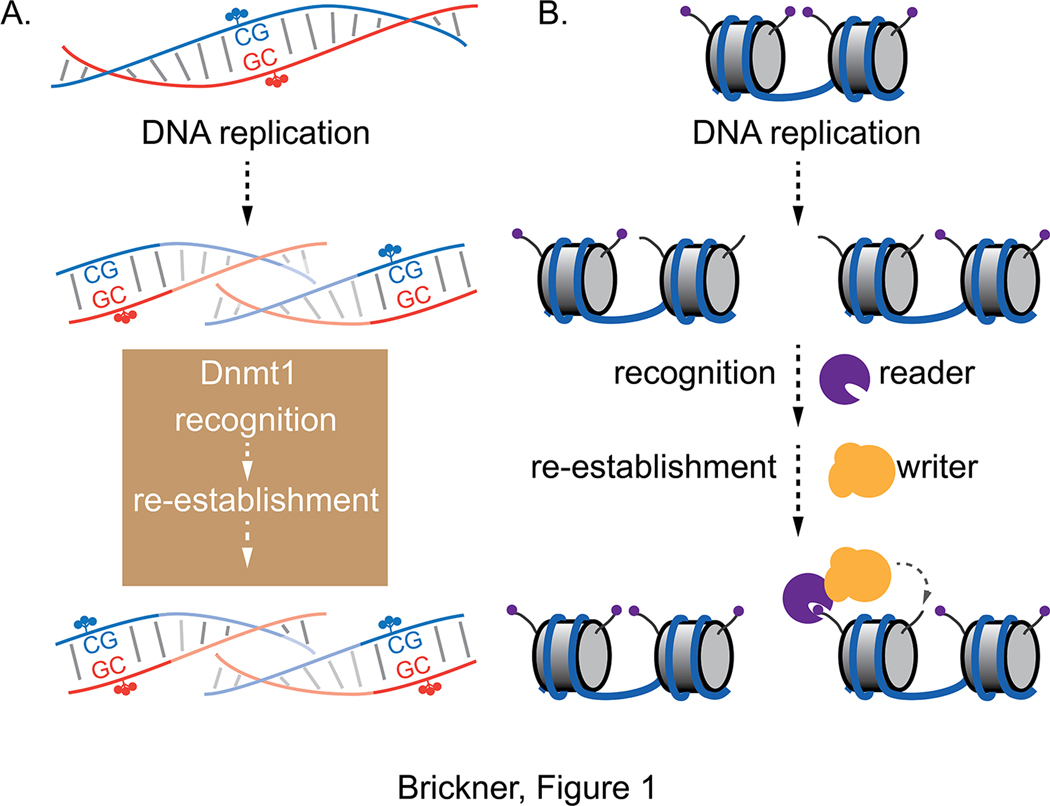
Figure 1. Read-write mechanisms for replicating chromatin marks.
A. Following DNA replication, hemi-methylated CpG sites are recognized by Dnmt1, which re-establishes symmetric methylation. This protein possesses the ability to both recognize the appropriate substrate and to catalyze the reaction. B. Because histone H3/H4 nucleosome cores are reincorporated nearby but randomly distributed between the daughter products of DNA replication, approximately half of the nucleosomes contain parental, local H3/H4 tetramers and half contain new H3/H4 tetramers. The parental nucleosomes can be recognized by reader proteins that bind to specific histone marks. Recognition by readers leads to recruitment of writers, which re-establish these marks on neighboring nucleosomes after DNA replication. C. Inheritance of H3K9 methylation. Both the writer Clr4/Suv29h and the reader Swi6/HP1 can bind H3K9me3. Because Swi6 physically interacts with Clr4, this provides both a direct and an indirect mechanism for Clr4 recruitment to H3K9me3-marked loci. Binding of H3K9me3 to Clr4 also stimulates catalytic activity of the enzyme. D. Inheritance of H3K27 methylation. The PRC2 Polycomb complex contains both a reader (EED; light green) and a writer (EZH2; dark green). Binding of EED to H3K27me3 both recruits PRC2 to chromosomal sites with this mark and stimulates EZH2 catalytic activity to promote methylation of lysine 27 on neighboring nucleosomes.