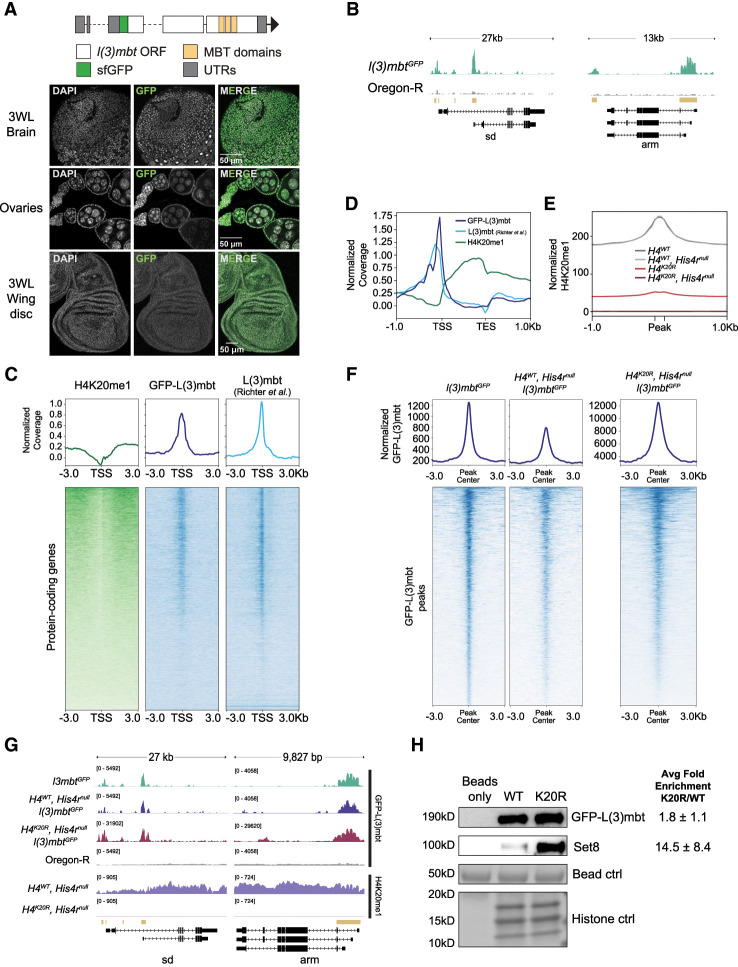
Figure 6.
L(3)mbt binds the genome independently of H4K20me. (A, top) Diagram of a GFP-tagged l(3)mbt allele generated using CRISPR. (Bottom) Confocal images of GFP-L(3)mbt accumulation in the nuclei of third instar larval brains, adult female ovaries, and third instar larval wing discs. (B) A representative locus depicting normalized GFP-L(3)mbt CUT&RUN coverage. Gold bars indicate peaks of GFP signal in l(3)mbtGFP (top) relative to Oregon-R control (bottom) as defined by merged 150 bp sliding windows. (C) Heat (bottom) and summary (top) metaplots of z-normalized wing disc H4K20me1 CUT&RUN, wing disc GFP-L(3)mbt CUT&RUN, and L(3)mbt ChIP-seq (Richter et al. 2011) signal at protein-coding genes. Plots are centered at the TSSs and flanked by 3 kb of unscaled sequence. Each row represents a single gene, and genes are ordered by mean H4K20me1. The summary plot depicts mean signal for H4K20me1 (green), GFP-L(3)mbt (blue), or L(3)mbt (light blue) at each position in the metaplot (50 bp bins). (D) Summary metaplot of z-normalized H4K20me1 (green), GFP-L(3)mbt CUT&RUN (blue), and L(3)mbt ChIP-seq (light blue) at high H4K20me1 genes. (E) Summary metaplot of spike-in-normalized H4K20me1 CUT&RUN coverage at H4K20me1 peaks in the indicated genotypes. Each peak is scaled to 200 bp and flanked by 1 kb of unscaled sequence. The summary plot depicts mean signal at each position in the metaplot (50 bp bins) for all peaks. (F) Summary (top) and heat (bottom) metaplots of spike-in-normalized GFP-L(3)mbt CUT&RUN signal. Plots are centered at the GFP-L(3)mbt peak, and each peak is flanked by 3 kb of unscaled sequence. Each row represents a single peak, and peaks are ordered by mean GFP-L(3)mbt in l(3)mbtGFP. (G) Representative locus depicting spike-in-normalized GFP-L(3)mbt and H4K20me1 CUT&RUN signal. Gold bars indicate peaks of GFP signal in l(3)mbtGFP relative to Oregon-R control as defined by merged 150 bp sliding windows. (H) Western blot using anti-GFP (top) or anti-Set8 (middle) antibodies following recombinant nucleosome binding assay with nuclear lysate from third instar larvae and either wild-type (WT) or H4K20R (K20R) recombinant nucleosomes. (Bottom) Histone proteins from Swift protein stain were used as loading control. Average binding of Set8 and GFP-L(3)mbt on K20R versus WT nucleosomes ±standard deviation from three biological replicates is shown at the right.