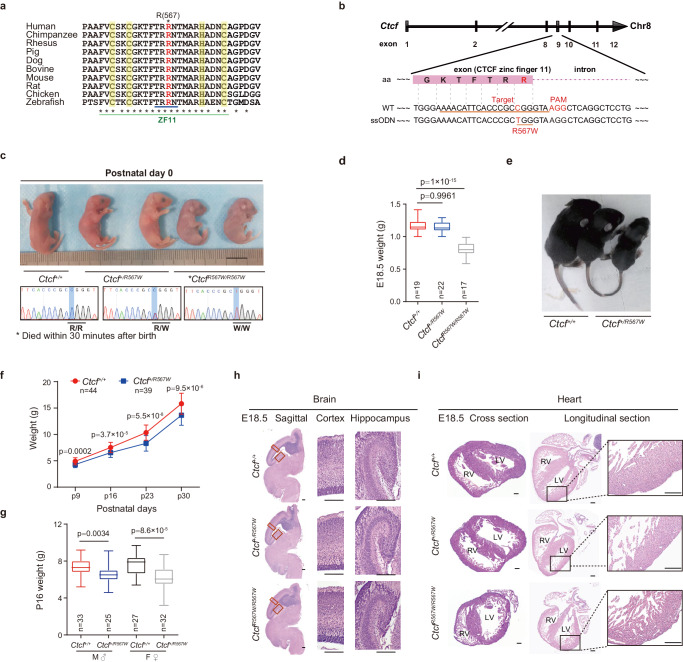
Fig. 1. Construction of CTCFR567W-mutant mice and phenotypic analysis.
a The amino acid (aa) conservation of CTCF around R567 across species. CTCF ZF11 residues annotated in UniProt are marked with a green line, DNA-binding residues (residues 565–568) with a blue line, and Zn2+ coordinating residues with a yellow band. R567 is highlighted in red. b CRISPR/Cas9 knock-in of the CTCFR567W mutation diagram. The sgRNA targeting sequence (Ctcf exon 9) and the mutation site R567W (R, Arg, CGG; W, Trp, TGG) are underlined. The mutated base and the PAM sequence NGG are in red. The aa sequence around the mutation site is highlighted with a pink box. c Representative photograph and Sanger sequencing results of neonatal mice. Scale bar, 10 mm. The mutation base C > T is highlighted in blue. d Box and whisker plots showing the body weight (95% confidence interval) of E18.5 mice (Ctcf+/+: n = 19, Ctcf+/R567W: n = 22, CtcfR567W/R567W: n = 17). e Two-week-old Ctcf+/+ and Ctcf+/R567W mice images. f Weight curves of postnatal day 9 (P9), P16, P23, and P30 Ctcf+/+ and Ctcf+/R567W mice (Ctcf+/+: n = 44, Ctcf+/R567W: n = 39). g Box and whisker plots showing the body weight (95% confidence interval) of P16 mice for different sexes of Ctcf+/+ and Ctcf+/R567W mice (Ctcf+/+: M, n = 33, F, n = 25, Ctcf+/R567W: M, n = 27, F, n = 32; M: male, F: female). h, i Sagittal brain and heart paraffin H&E staining images of Ctcf+/+, Ctcf+/R567W, and CtcfR567W/R567W mice at E18.5. Enlarged views of the cortex, hippocampus, and left ventricular wall are shown on the right. Scale bars, 200 μm. Quantitative data are presented as mean ± SD. p-values were obtained by one-way ANOVA with Dunnett’s multiple comparisons test adjustment (d), two-way ANOVA with Šídák’s multiple comparisons test adjustment (f), and two-tailed unpaired t-test (g) are indicated in the graphs. Experiments in (h, i) were repeated independently three times with similar results. All Box and whisker plots show lower and upper quartiles (box limits), median (center line) and minimum to maximum values (whiskers). Source data are provided as a Source Data file.