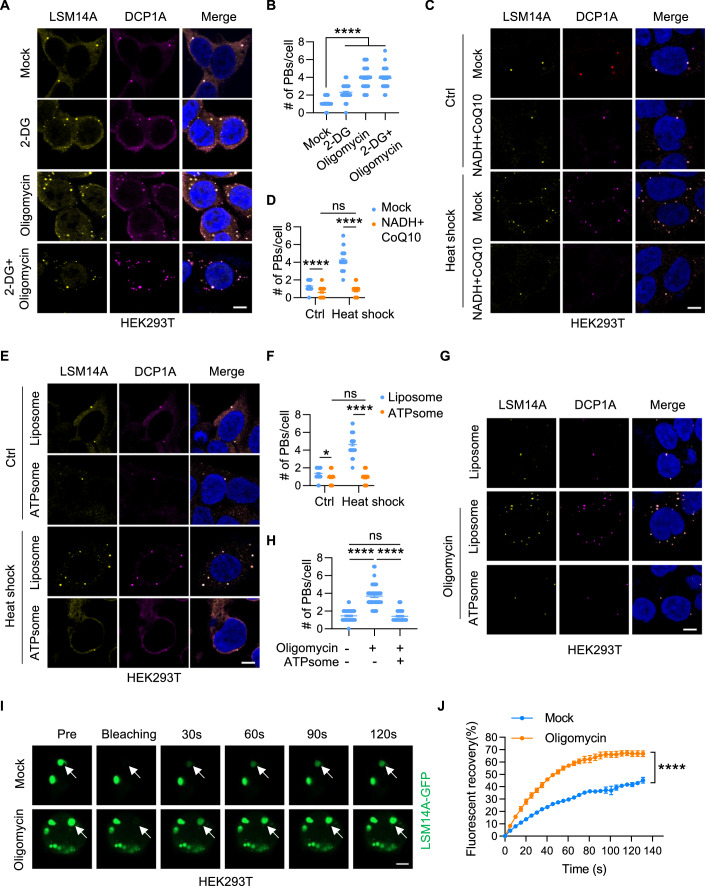
Figure 1. ATP depletion fosters P-bodies formation.
(A, B) HEK293T cells were treated with 2-DG or/and oligomycin (2 h) and stained for LSM14A and DCP1A. Representative images are shown in (A). The number of P-bodies within each cell from one of three biological replicates is plotted in (B) (n = 50). Scale bar, 5 μm. Error bars indicate SEM. ****P < 0.0001 (Student’s t test). (C, D) HEK293T cells treated with or without NADH+CoQ10 (2 h) were exposed to heat shock (42 °C) (1 h) and stained for LSM14A and DCP1A. Cells were imaged (C), and the number of P-bodies within each cell was quantified (D) (n = 50). Scale bar, 5 μm. Error bars indicate SEM. ns: no significance, ****P < 0.0001 (two-way ANOVA). (E, F) HEK293T cells treated with liposome or ATPsome (1 h) were exposed to heat shock (42 °C) and stained for LSM14A and DCP1A. Representative images are shown in (E). The number of P-bodies within each cell is plotted in (F) (n = 50). Scale bar, 5 μm. Error bars indicate SEM. *P < 0.05, ****P < 0.0001 (two-way ANOVA). (G, H) HEK293T cells subjected to different treatments were stained for LSM14A and DCP1A. Cells were imaged (G), and the number of P-bodies within each cell was quantified (H) (n = 50). Scale bar, 5 μm. Error bars indicate SEM. ns: no significance, ****P < 0.0001 (Student’s t test). (I, J) HEK293T cells expressing LSM14A-GFP were treated with DMSO and oligomycin, and the relative mobility of LSM14A-GFP was determined by FRAP (I, J). White arrows indicate the target droplets for FRAP. Scale bar, 5 μm. n = 3 experiments, Error bars indicate SEM. ****P < 0.0001 (two-way ANOVA). Source data are available online for this figure.