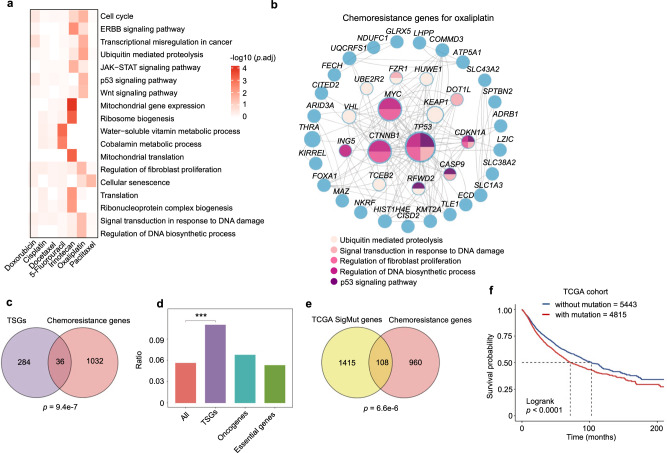
Fig. 2. A systematic view on chemoresistance genes.
a Enriched functional terms from Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) for chemoresistance genes of indicated chemotherapy drugs. Unpaired two-sided t test for p value with Benjamini-Hochberg (BH) adjustment. b Protein-protein interaction network for chemoresistance genes of oxaliplatin. The genes belonging to indicated functional terms are color-coded. c Venn diagram showing the overlap between tumor suppressor genes (TSGs) and chemoresistance genes. The overlap is significant as calculated by the Hypergeometric test (two-sided, p = 9.4e-7). d Enrichment ratio of chemoresistance genes in all genes, TSGs, oncogenes, and essential genes. Unpaired two-sided t test, p = 6.3e-4. e Overlap of significantly mutated genes from The Cancer Genome Atlas (TCGA) cohorts (pan-cancer) and chemoresistance genes identified by our screens. The overlap is significant as calculated by the Hypergeometric test (two-sided, p = 6.6e-6). f Overall survival analysis of all TCGA patients using the mutational status of 108 overlapping genes in e. Kaplan–Meier statistical test, p = 2.3e-9.