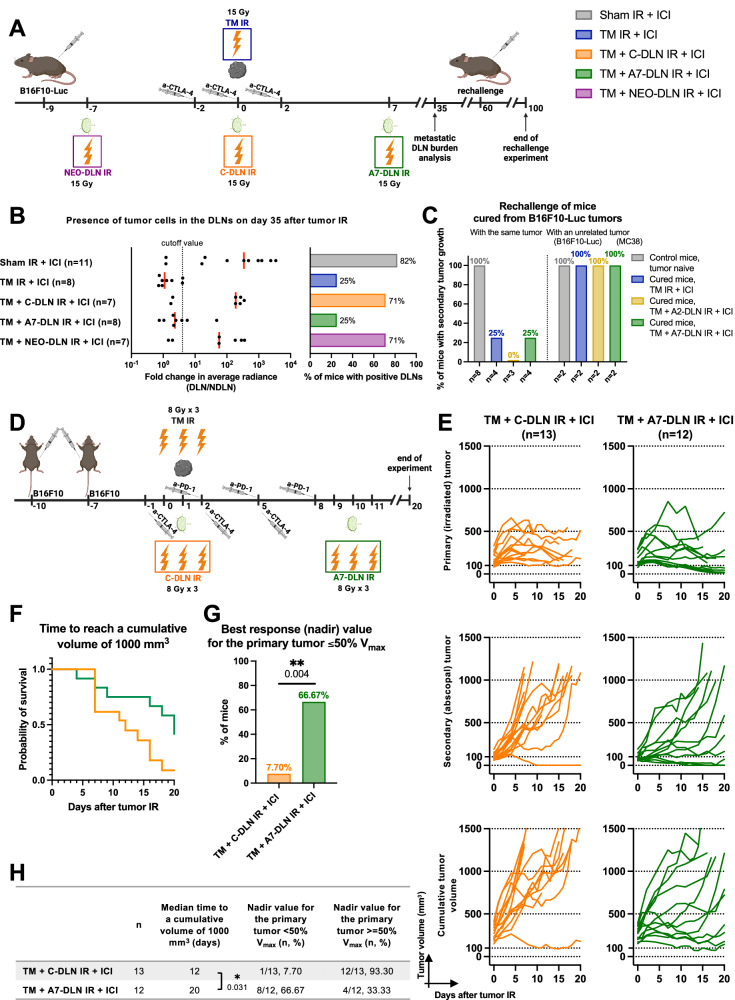
Fig. 6. Adjuvant draining lymph node irradiation allows for the development of regional control and the induction of long-lasting tumor-specific immunity.
A B16F10-Luc tumor-bearing mice received α-CTLA-4 and tumor IR, with or without DLN IR at different timepoints, as illustrated. For the evaluation of the presence of tumor cells, DLNs were excised on day 35 after tumor IR. For the rechallenge, cured mice were injected on the contralateral flank, using either the same B16F10-Luc or antigenically unrelated MC38 cells on day 60 after tumor IR. Tumor growth was followed for 40 days after the rechallenge. B Quantification of tumor cell positivity in the DLNs. Left: Fold change in the average radiance of the DLN compared to the contralateral NDLN. Dotted line indicates the cutoff value for tumor cell positivity, defined as a 300% increase in the signal over the NDLN (fold change >4). Each dot represents an individual mouse. Red line represents the median. Right: Quantification of mice with DLN metastasis, using the cutoff value of 4. Number of mice in each group is indicated in the graph. C Percentage of mice with second tumor growth after rechallenging, using either the same B16F10-Luc or antigenically unrelated MC38 cells. Number of mice in each group is indicated in the graph. D Mice bearing two B16F10-Luc tumors received α-CTLA-4, α-PD-1 and tumor IR, as indicated. DLN IR was performed either concomitantly (C-DLN IR, orange), or 7 days after tumor IR (A7-DLN IR, green). All targets received 8 Gy per fraction in three fractions. Tumor growth was followed over 20 days. Treatment response represented by individual tumor growth curves (E), Kaplan–Meier survival analysis (F and H), and the percentage of mice achieving a 50% or more decrease in the primary (irradiated) tumor volume (i.e. nadir value ≤ 50% Vmax) (G and H). Time to reach a cumulative volume (i.e. the sum of the primary and secondary tumor volume on a given day) of 1000 mm3 was used as the endpoint for Kaplan–Meier analysis. Each line in E represents an individual mouse. Number of mice in each group is indicated in E. Logrank test (Mantel–Cox) was used to compare the survival curves; corresponding p values are displayed in H. Two-sided Fisher’s exact test was used to compare the categorical data in G. All p values are displayed, with *, ** and *** indicating p < 0.05, p < 0.01 and p < 0.001, respectively. Source data are provided as a Source Data file. Figures A and D, created with BioRender.com, released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license.