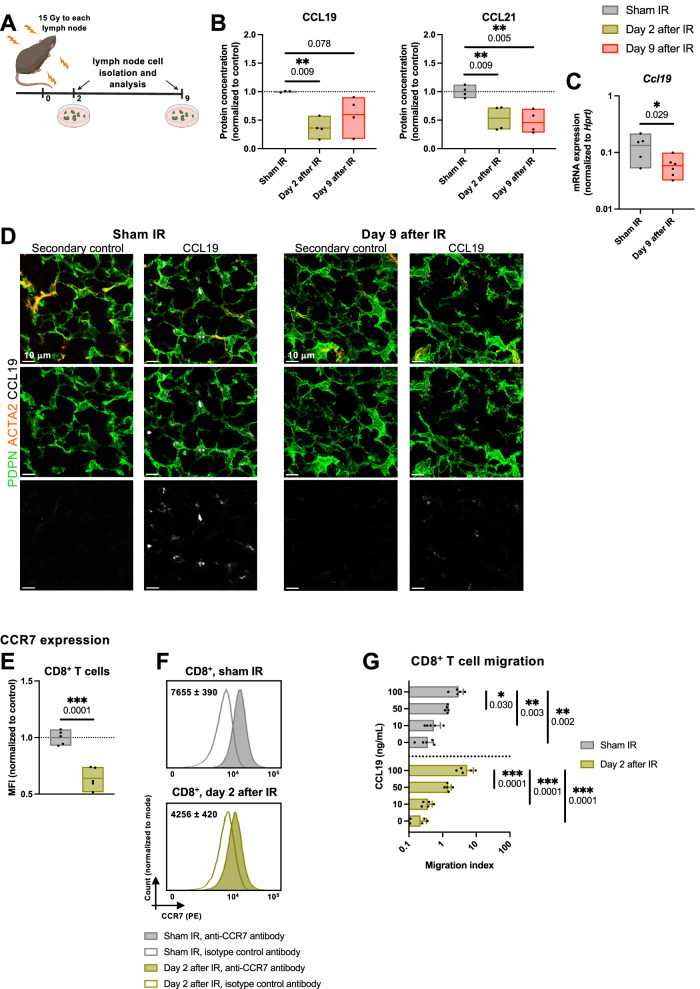
Fig. 9. Lymph node irradiation interferes with the CCR7-CCL19/CCL21 immune cell homing axis.
A Brachial, axillary, and inguinal lymph nodes on both sides of healthy C57BL/6 mice were irradiated using 15 Gy. Lymph nodes were harvested on days 2 (green) and 9 (red) after IR. B CCL19 and CCL21 protein concentration in the DLNs, expressed relative to the average value of the sham-irradiated mice (gray). n = 4 mice per group. C Ccl19 mRNA expression in fibroblastic reticular cells, normalized to Hprt. n = 6 mice per group. D Fluorescence microscopy of a section of a sham-irradiated (left) and an irradiated inguinal lymph node (right), on day 9 after IR. Sections are stained for podoplanin (PDPN) (green), actin alpha 2 (ACTA2) (orange) and CCL19 (white). Representative sections from n = 2 mice per treatment group, 2 lymph nodes per mouse. E, F CCR7 expression on CD8+ T cells. E Geometric mean of the fluorescence intensity (MFI) of CCR7 on CD8+ T cells, normalized to the corresponding average MFI value of in the sham-irradiated group. F Representative histograms. Values in histograms indicate the average MFI of CCR7 on CD8+ T cells (shaded histograms) minus the MFI of the corresponding isotype control (transparent histograms) ± standard deviation (SD). n = 5 mice per group. G Transwell migration assay of CD8+ T cells isolated from sham-irradiated lymph nodes (gray) and lymph nodes irradiated with 15 Gy 2 days prior to resection (green). Migration index is calculated by dividing the number of migrated cells in the given condition with the basal migration value (i.e. the number of migrated cells towards the bottom chamber containing 10% FBS). n = 5 mice per group. Each dot represents an individual mouse. Floating bars in B, C and E span from the minimal to the maximal value of each group. Line indicates the mean. Bar width in G represents the mean value, with error bars indicating the SD. According to the Shapiro–Wilk test, all data followed a normal distribution. Groups were compared using the two-sided unpaired t test (C and E), one-way ANOVA with Holm–Sidak’s multiple comparisons test (B) and two-way ANOVA with Holm–Sidak’s multiple comparisons test (G). All p values are displayed, with *, ** and *** indicating p < 0.05, p < 0.01 and p < 0.001, respectively. Source data are provided as a Source Data file. Figure A, created with BioRender.com, released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license.