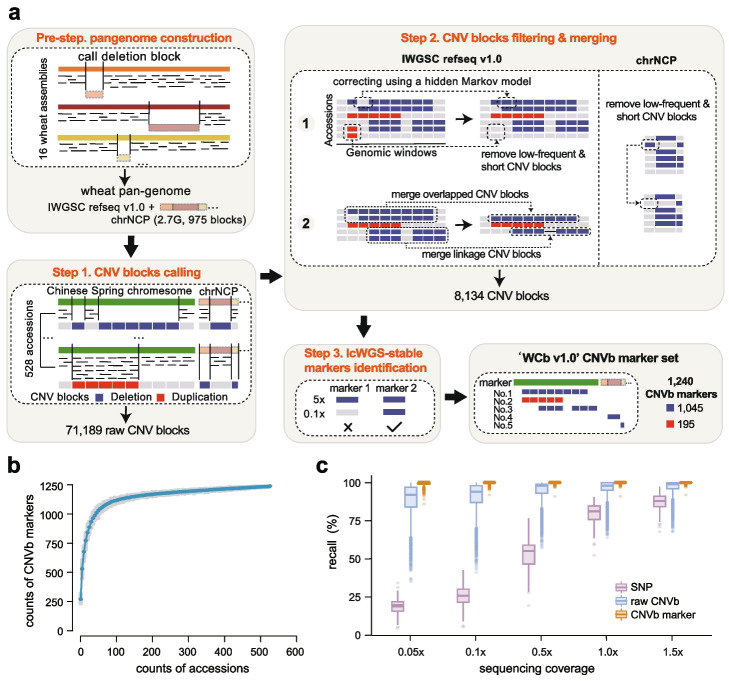
Fig. 2.
Development and evaluation of CNVb markers. a Pipeline to identify CNVb markers against wheat pan-genome. Pre-step, the pan-genome was constructed by combining the Chinese Spring assembly and the unmapped blocks of Chinese Spring relative to the other 16 wheat assemblies. Step 1, the initial CNV blocks of 528 high-quality wheat resequencing accessions were identified with a 100 Kb window. Step 2, a hidden Markov model (HMM) was introduced to smooth noisy signals, and then low frequency and short CNV blocks were filtered from the retained CNV blocks. CNV blocks with reciprocally overlapped regions larger than 80% were merged as a single CNVb cluster, and linkage clusters with close distances were further combined. Step 3, the final CNVb markers were extracted by eliminating those with low recall rates identified by ultra-low-coverage whole genome sequencing (ulcWGS), using CNVb markers identified by high-coverage whole genome sequencing as the ground truth. b Saturation analysis of CNVb markers. Five accessions were randomly added each time. The shaded area represents 100 replications for each sampling. The blue dot represents the average number of CNVb markers across 100 repetitions. c Comparison of the accuracy of SNPs, raw CNVs, and CNVb markers identified at low sequencing depth