Abstract
Cell migration is the major driver of invasion and metastasis during cancer progression. For cells to migrate, they utilize the actin–myosin cytoskeleton and adhesion molecules, such as integrins and CD44, to generate traction forces in their environment. CD44 primarily binds to hyaluronic acid (HA) and integrins primarily bind to extracellular matrix (ECM) proteins such as collagen. However, the role of CD44 under integrin-mediated conditions and vice versa is not well known. Here, we performed traction force microscopy (TFM) on U251 cells seeded on collagen I-coated polyacrylamide gels to assess the functional mechanical relationship between integrins and CD44. Performing TFM on integrin-mediated adhesion conditions, i.e., collagen, we found that CD44KO U251 cells exerted more traction force than wild-type (WT) U251 cells. Furthermore, untreated WT and CD44-blocked WT exhibited comparable results. Conversely, in CD44-mediated adhesive conditions, integrin-blocked WT cells exerted a higher traction force than untreated WT cells. Our data suggest that CD44 and integrins have a mutually antagonistic relationship where one receptor represses the other's ability to generate traction force on its cognate substrate.
INTRODUCTION
Aggressive cell migration leads to tissue invasion and metastasis, one of the fundamental hallmarks of cancer, and is partly responsible for the severity of the disease.1 In glioblastoma, tumor invasion increases brain inflammation, which further promotes GBM progression.2 One of the main drivers of metastasis is cell migration, and understanding how cancer cells adhere to and migrate has been a major goal of cancer research. Cell migration is also vital to normal immune responses and developmental processes. During migration, transmembrane receptors, such as integrins and CD44, function as “clutches” to mechanically link the extracellular matrix (ECM) to the actin cytoskeleton, providing the cell with adhesion.3,4 Proteins such as talin and vinculin function as adaptors, mechanically linking the integrins to the actin cytoskeleton on integrin-mediated surfaces. At the same time, non-muscle myosin II works with F-actin to function as a “motor” by generating the force for the clutches to “pull” the cell body forward while the trailing edge of the cell detaches as the cell is pulled forward.3–5 While adhered, cells generate inwardly directed traction forces by pulling on their adhesive attachments to the local ECM.
According to previous models, the number of motors and clutches are major determinants of whether a cell can migrate efficiently in its environment via a motor-clutch mechanism.4 When motors are much higher than clutches, then cells exhibit low adhesion phenotype with cells that are morphologically circular and small in projected area. This phenotype is referred to as the cells being in a free-flowing state, where the excessive number of motors overpower and prevent the smaller number of clutches to maintain “grip” on the substrate.4 Although this low adhesion results in a low traction force, it does not necessarily result in diminished migration. In fact, previous studies have shown certain types of cancer cells in confined, low adhesion conditions can exert fast amoeboid-like migration.6 On the other extreme, when the number of clutches is much higher than the number of motors, the cells express a well adhered and well spread phenotype. These cells are referred to as being in a stalled state.4 The cell is well spread due to the strong adhesion, but since the motor number is low, it is difficult for the cell to generate sufficient force to detach protrusions on the trailing side of the cell, and thereby pull the cell, which results in inefficient migration but moderate to strong traction force.3 Although traction force does not have a direct correlation with migration, they are mechanistically related to one another. Therapeutically, we seek to reduce cancer cell migration without significantly impairing antitumoral immune cell migration, and thereby increase patient survival.
Integrins are one of the two major clutches involved in glioblastoma migration. They are heterodimeric transmembrane receptors that play key roles in several other cell processes such as mediating cell adhesion and signaling.7 Activation is required for integrins to bind to their ECM ligand.8 Once integrins bind to the ECM, they form focal adhesion complexes consisting of several proteins, such as vinculin, α-actinin, and other components.9 Focal adhesion formation begins as the head domain of talin binds to the cytoplasmic tail of β-integrin.10,11 There are over 50 different cytoskeletal proteins that bind to the focal adhesion complexes, and they cluster over ECM bound integrins to connect the clutches to the cytoskeleton.12 In these clusters, there is a combination of specialized proteins, such as adaptors, signaling, regulatory, and mechanosensing proteins, that work together to enable cells to detect the mechanical properties of their environment.13 Integrins function in cell signaling by communicating intracellularly via FAK activated pathways.14 Outside the cell, integrins can bind to distinct ECM proteins based on their αβ-heterodimeric subunits.15 Once the extracellular domain (ECD) of the integrin binds to its cognate ECM ligand, the intracellular domain (ICD) attaches to cytoskeletal proteins, such as talin and vinculin,16,17 which serve as adaptors for connecting integrins to F-actin. Through this mechanical linkage, non-muscle myosin II pulls the F-actin to generate a retrograde pulling force on the ECM.12
The other major clutch in glioblastoma is the transmembrane receptor CD44, which is overexpressed in multiple cancers.18–20 One of its main functions is to bind via its ECD to hyaluronic acid (HA), although previous studies have reported interactions with collagen, laminin, and several growth factors.21,22 Via its ICD, CD44 engages in signaling pathways, including Ras and PI3K, which downstream leads to increased proliferation, adhesion, and migration in glioblastoma.23 Like integrins, once CD44 binds to its ligand, ICD-CD44 binds to adaptor proteins belonging to the ERM family of proteins, consisting of ezrin, radixin, and moesin.24 These adaptors connect the CD44 to F-actin, which is functionally akin to how talin and vinculin act as adaptors to connect integrins to F-actin, where it can generate forces to enable cell movement.24
The motor-clutch model predicts that traction force increases as the number of clutches increases, then once the cell enters a stalled state, the traction force plateaus.3 However, having an increase in clutches does not directly lead to increased migration. In fact, cells must have a balanced system of motors and clutches for optimal cell migration.25–27 The environment also plays a vital role on cell adhesion and migration since cells can use mechanosensing to adapt to their environment.28 The number of motors and clutches could also be adjusted to match the environment.27 In a previous study, optimal migration of U251 cells peaked at a lower stiffness when treated with blebbistatin and cycloRGD, drugs which decrease the number of motors and clutches, respectively.27 A change in the environment could lead to a change in motor and clutch number. Some factors, such as the stiffness of the substrate, concentration of ECM, and type of ECM, also contribute to migration efficiency. For U87 cells, and several other glioblastoma cell lines, cell migration peaks at an intermediate Young's modulus.29 The motor-clutch model also predicts that there is an optimal environmental stiffness at which cell migration is maximal, which was observed for several cell types, including U251 glioblastoma cells.27,30 Finally, the ECM type plays a role in determining the effective number of clutches because of adhesion receptors distinct ligand compatibilities. For example, collagen interacts with several integrin subtypes, meanwhile hyaluronic acid exclusively binds to CD44.31,32 These two clutches work together presumably simultaneously in an in vivo environment during cell adhesion and migration. A previous study suggested that integrins and CD44 use signaling pathways to coordinate traction via crosstalk.33 In another study, CD44 stimulation increased integrin-mediated cell adhesion via cell signaling.34
TFM works by tracking fluorescent beads embedded in an elastic gel, these beads are used as displacement markers. Polyacrylamide gels (PAGs) are coated with collagen I for integrin-mediated ECM or a monoclonal antibody for CD44.35 As the cell adheres to the PAG and exerts cell traction, it establishes adhesions, which deform the gel and allow us to calculate traction strain energy or force.36
In this study, we use computational modeling, single cell migration assays, and traction force microscopy (TFM) to observe cell traction, as well as cell migration and morphology, to assess the functional mechanical relationship between integrins and CD44 on their mediated substrate. We assess the extent to which CD44 and integrins are independent, or whether they can communicate and work synergistically or antagonistically to mediate cell traction.
RESULTS
CD44 inhibition enhances traction force on collagen-mediated conditions
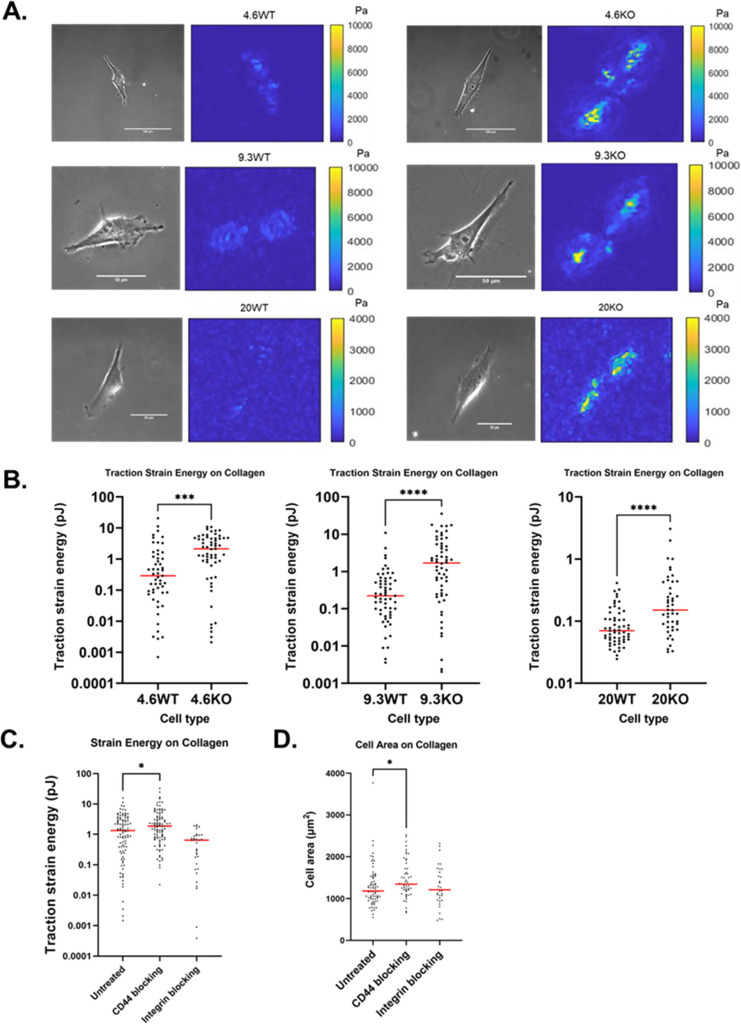
To assess the influence of CD44 on integrin-mediated adhesion to collagen, we plated U251 wild-type (WT) and CD44 knockout (CD44KO) on collagen I-coated polyacrylamide hydrogels across a range of substrate stiffnesses (4.6 kPa, 9.3, and 20 kPa Young's modulus). Since CD44 has a limited ability to adhere to collagen, we expected to see no difference in traction force between U251WT (WT) and CD44KO (KO) cells. Contrary to expectation, we found that although WT and CD44KO cells appear morphologically similar and spread to similar extents, CD44KO cells exert sevenfold more traction force, quantified as traction stain energy, on 4.6 and 9.3 kPa [Fig. 1(a); p = 0.0001 and p < 0.0001, respectively] and twofold more traction force on 20 kPa [Fig. 1(a): p < 0.0001]. Compared to WT cells, CD44KO cells exert greater traction strain energy on collagen across the range of stiffness from 4.6 to 20 kPa [Fig. 1(b)]. On 4.6 and 9.3 kPa, traction strain energy is similar, but when stiffness increased to 20 kPa, we observed traction strain energy decreases in both cell types, consistent with previous results that showed an optimal stiffness for U251 traction strain energy on collagen-coated PAGs at 4.6–9.3 kPa.27
FIG. 1.
CD44KO cells exert more traction force than WT cells on collagen-coated surfaces. (a) Traction stress magnitude map comparison of WT and CD44KO on 4.6 kPa collagen-coated gel. In both cases, a phase contrast image of the cell is on the left and traction stress map is on the right. (b) Traction strain energy of WT and CD44KO as a function of substrate stiffness. For 4.6 kPa: n = 53 on WT, n = 59 on CD44KO; for 9.3 kPa: n = 59 on WT, n = 60 on CD44KO; and for 20 kPa: n = 59 on WT, n = 46 on CD44KO. (c) Traction strain energy of WT untreated, CD44 blocked, and integrin blocked on 4.6 kPa collagen. For untreated, CD44 blocked, integrin blocked on collagen, n = 96, 97, and 33, respectively. (d) Cell area of WT untreated, CD44 blocked, and integrin blocked on 4.6 kPa collagen. The red line shows the median. P calculated by the Mann–Whitney test. * denotes P < 0.05, ** denotes P < 0.01, *** denotes P < 0.001, and **** denotes P < 0.0001.
To further test whether reduction in CD44 activity causes traction force to increase on collagen-coated surfaces, we used a monoclonal anti-CD44 antibody known to block CD44 adhesion (IM7).33 When blocking CD44 expressed by WT cells, we again observed an increase (p = 0.03) in cell traction force in CD44-blocked cells compared to control untreated cells [Fig. 1(c)]. As a positive control, anti-integrin antibodies were used to block integrins from binding to the surface and confirm that decreasing integrins on collagen-coated substrates reduces traction force. Two integrin monoclonal antibodies were used simultaneously to block integrins from collagen; one antibody targeted the αV subunit (SC56-07), and the second antibody targeted the α3 subunit (P1B5). These two integrins were targeted for blocking because they had the highest mRNA expression of all the αintegrin subunits in U251 cells in our previous study.27 The cell area in CD44-blocked WT cells had an approximate 12% increase in cell area compared to untreated WT [Fig. 1(d), p = 0.0085]. We conclude that CD44 acts antagonistically to suppress integrin-mediated traction forces on collagen-coated surfaces.
Blocking integrins results in a higher traction force via CD44-mediated adhesion
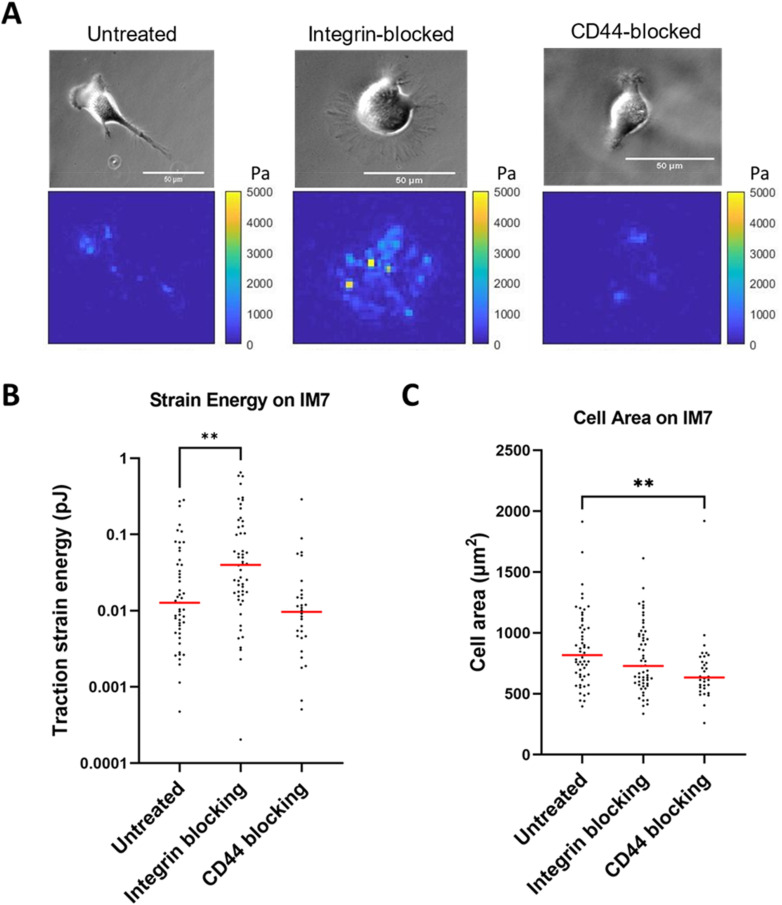
We hypothesized that if CD44 is antagonizing integrin-mediated traction, then perhaps there might be reciprocal crosstalk that causes integrins to antagonize CD44-mediated traction. Thus, we tested WT cells under the following treatments: untreated, CD44 blocking, or integrin blocking, but this time under CD44-mediated substrate conditions. We coated PAG with a CD44-specific monoclonal antibody (clone IM7) to generate a condition where only CD44 clutches should mediate adhesion and traction force. The same antibody used for ECM coating was added to the medium as a control. Under CD44-mediated adhesion conditions, the median traction force for untreated, integrin-blocked, and CD44-blocked WT was 0.013, 0.040, and 0.010 pJ, respectively [Fig. 2(a)]. We found that integrin-blocked cells exert threefold greater traction strain on IM7-coated substrates than control cells [Fig. 2(a); p = 0.0016]. When we measured cell area on the IM7-coated substrate, we found that untreated and integrin-blocked cells are not different [Fig. 2(b); p = 0.17]. As expected, the cell area observed with CD44-blocked cells is smaller than untreated [Fig. 2(b); p = 0.0016]. However, CD44-blocked cells on a CD44-mediated surface resulted in reduced adhesion, which resulted in a bias where we can only observe the cells within the population with the strongest adhesions. We believe cells are using CD44 exclusively to interact with the IM7-coated PAGs. We found that the majority of U251 cells did not adhere to IM7-coated gels when CD44 is blocked via soluble IM7. Since U251 WT cells have heterogeneous CD44 expression, we surmised that the minority of cells that still adhered when measuring traction strain energy [Fig. 2(b)] were exhibiting a relatively higher amount of CD44, enabling them to resist detachment in the presence of soluble IM7. In Fig. 2(c), we show a decrease in the cell area in CD44-blocked cells, suggesting less adhesion. The simplest explanation is that these cells are significantly, but not entirely, inhibited by soluble IM7, and that they still use a few remaining unblocked CD44 molecules to adhere to the IM7-coated gels as it is difficult to achieve 100% inhibition via addition of an antibody. Although a larger cell spread area generally means more traction strain energy, since larger cells have more adhesive clutches to bind the substrate surface, the cell area does not necessarily correlate with traction force [Figs. 2(a) and 2(b)]. The results provide evidence for antagonism of CD44-mediated traction forces via integrins.
FIG. 2.
Integrin-blocked WT cells exert more traction force than untreated WT cells on anti-CD44 antibody-coated surfaces. (a) Traction force for untreated, integrin-blocked, and CD44-blocked U251 cells on IM7-coated PAGs, which supports CD44-mediated adhesion. (b) Cell area was relatively unaffected by integrin blocking and reduced in CD44-blocked conditions on IM7-coated PAG. For untreated, CD44 blocked, integrin blocked on collagen, n = 49, 54, and 30, respectively. (c) Transmitted images (top) and traction maps (bottom) of each condition. The red line shows median. P value was calculated by the Mann–Whitney test. ** denotes P < 0.01.
Migration is unaffected by CD44-KO on collagen-coated surfaces
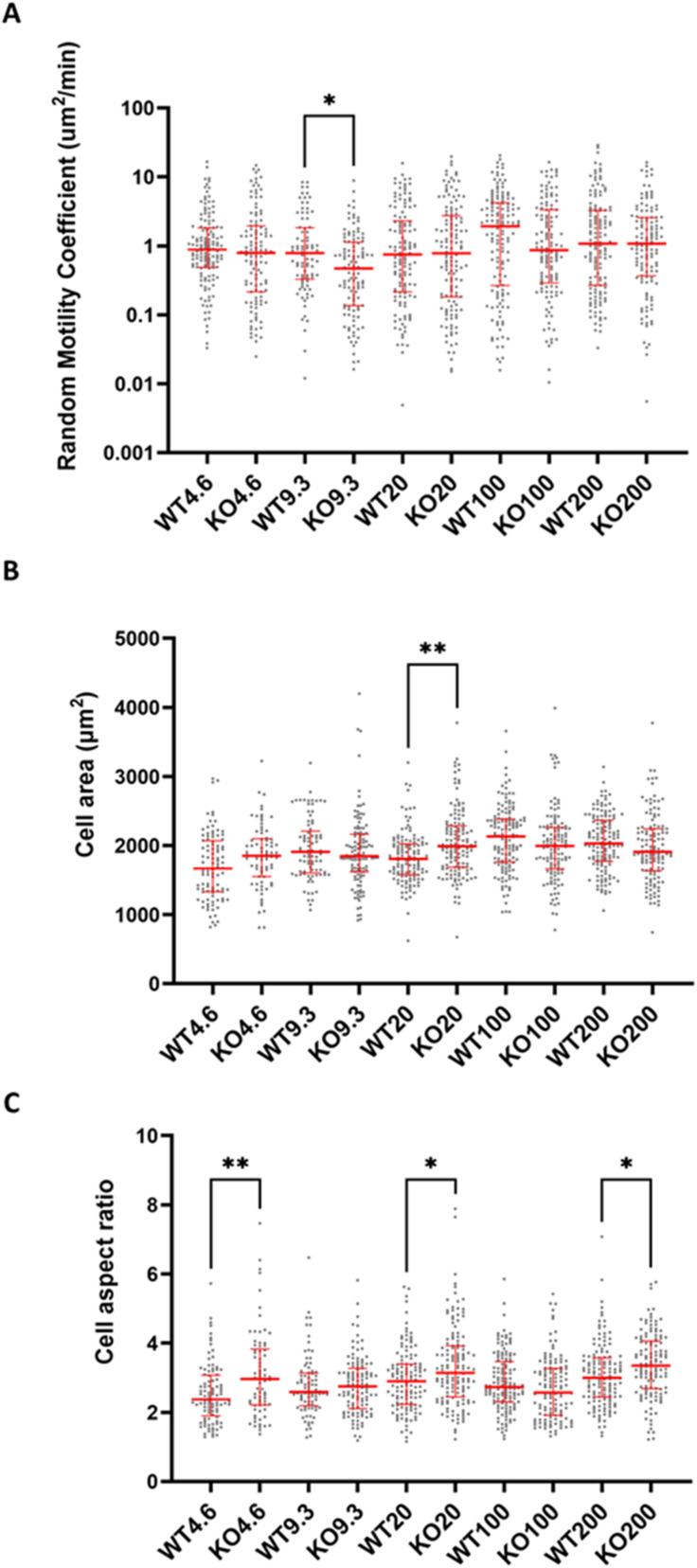
Since knocking out or blocking CD44 potentiated traction force on collagen-coated surfaces, i.e., integrin-mediated adhesion substrate, we were interested in assessing whether cell migration was affected as well. We measured cell migration for WT and CD44KO U251 cells on collagen-coated PAGs with stiffnesses ranging from 4.6 to 200 kPa (see Fig. S1 for confirmation of CD44 knockout). On all stiffnesses except for 9.3 kPa, we observed no difference in migration between WT and CD44KO [Fig. 3(a)]. At 9.3 kPa, WT and CD44KO cells had a median random motility coefficient (RMC) of 0.76 and 0.47 μm2/min, respectively (P = 0.027). WT migration appeared to peak at 100 kPa with a random motility coefficient of 1.9 μm2/min, similar to what was observed in our previous studies of U251 WT cells on collagen-coated surfaces;27 however, there is no peak for the CD44KO cells.
FIG. 3.
Migration of WT and CD44KO in integrin-mediated conditions is similar across a range of stiffnesses. (a) Migration, (b) cell area, and (c) cell aspect ratio across stiffnesses (Young's modulus, unit kPa) for WT and CD44KO, respectively. Starting from 4.6 kPa and increasing stiffness, n = 140, 85, 130, 142, and 139 for WT and 115, 115, 137, 125, and 127 for CD44KO. Statistical significance was analyzed between WT and CD44KO on each stiffness. The red line shows the median along with the interquartile range. P values were calculated by the Kruskal–Wallis test. * denotes P < 0.05 and ** denotes P < 0.01.
In addition to cell migration, cell spread area was also quantified to further assess potential antagonism between CD44 and integrins. Since a larger cell area suggests a more adhesive cell, we expected to see a larger cell area in CD44KO. However, when comparing WT and CD44KO across stiffnesses, we found that the cell area is larger for CD44KO on only one stiffness [Fig. 3(b); P = 0.0011]. Finally, the cell aspect ratio serves as a measure of cell polarization, with a higher cell aspect ratio tending to correlate with cell area and migration.27 CD44KO has a higher cell aspect ratio on three stiffnesses: 4.6, 20, and 200 kPa [Fig. 3(c); P = 0.0044, 0.05, and 0.036, respectively].
Modeling the mechanics of mutual antagonism between CD44 and integrins
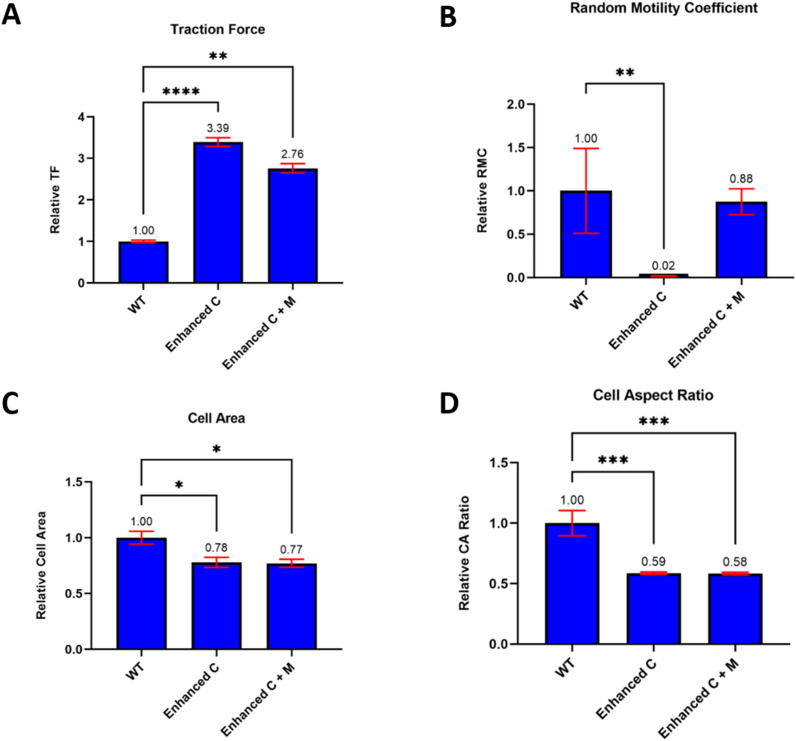
To explain these experimental observations, we used a motor-clutch-based cell migration simulator, as previously described.27 In our experimental data, we found an increase in traction in CD44KO cells on collagen I-coated PAGs, but no notable change in migration, spread area, or aspect ratio. Here, we examined three scenarios: (1) WT (control), (2) CD44KO with an increased number of clutches only (enhanced C), and (3) CD44KO with an increased number of clutches and motors (enhanced C + M). WT cells were simulated with 1000 motors and 750 clutches.27 The other two cases represent two possibilities of what is happening in the CD44KO cells on collagen-coated surfaces. In one scenario for CD44KO (C only), we simulate a “clutches only increase” scenario by assuming 1000 motors and 3000 clutches, which is expected to increase traction force by shifting the cell into a “stalled” state where traction forces are high and F-actin retrograde flow is nearly zero. For the other CD44KO scenario (C + M), the clutches and motors increase, which was simulated by 3000 motors and 2250 clutches, i.e., where both motors and clutches increase threefold. In all conditions cells were simulated on a stiffness of 100 pN/nm, comparable to a Young's modulus of 100 kPa for micrometer-scale adhesions. Compared to the WT control, traction force increased significantly in both CD44KO scenarios. The enhanced C scenario exerted a traction force that was increased 3.4-fold relative to WT, and in the enhanced C + M scenario, traction force was increased 2.7-fold [Fig. 4(a); P < 0.0001 and 0.0096, respectively]. Both scenarios are consistent with the experimentally observed increased traction force on collagen when CD44 is KO/inhibited and on α-CD44Ab-coated (IM7) surfaces when integrins are inhibited. However, in the clutches only scenario, the simulated cells exhibited a large drop in RMC that was not observed experimentally, down 50-fold relative to WT (P = 0.0016). In contrast, the enhanced motors and clutches showed no significant change, as observed experimentally [Fig. 4(b); P = 0.72]. Cell spread area was similar in both CD44KO scenarios. Cell area decreased by ∼22% for both CD44KO scenarios, i.e., enhanced clutches and enhanced motors and clutches [Fig. 4(c); P = 0.012 and 0.012, respectively]. Finally, cell aspect ratio decreased by ∼41% in both CD44KO scenarios, enhanced clutches and enhanced motors and clutches [Fig. 4(d); P = 0.0006 and 0.0003, respectively]. We conclude that the increased clutch and motor scenario provides a better description of the CD44KO compared to WT than the clutch-only scenario because it yields markedly increased traction strain energy with only modest effects on cell migration, area, and aspect ratio, as observed experimentally.
FIG. 4.
Cell migration simulator suggests that both motors and clutches are increasing upon CD44KO on collagen-coated surfaces. (a) Traction force, (b) random motility coefficient, (c) cell area, and (d) cell aspect ratio were modeled using the CMS. The data were normalized to WT control to focus on changes between WT and enhanced conditions. Results are given as mean +/- s.e.m. (N = 10 cells simulated per condition). P values were calculated by the Kruskal–Wallis test * denotes P < 0.05, ** denotes P < 0.01, *** denotes P < 0.001, and **** denotes P < 0.0001.
mRNA sequencing shows several gene expression alterations in CD44 knockout cells linked to cell adhesion and pathways
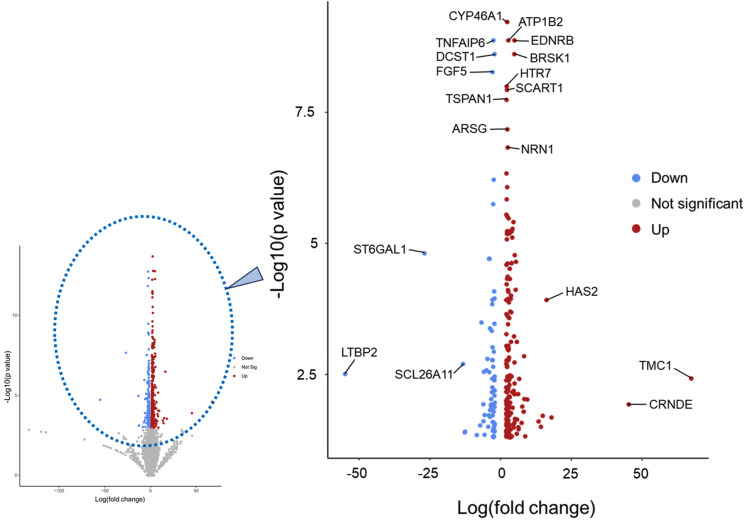
Next, we compared CD44 knockout and WT mRNA expression to see if we could find evidence for motor and clutch increase, as predicted in our model's simulations and experimental data. We do this by focusing on genes associated with integrin-mediated and CD44-mediated adhesion and migration. We expected to see upregulation of mRNA expression in integrin-associated genes and motor genes in CD44 knockout cells. Over 270 genes showed a significant fold change in expression (FC > 2, FDR < 0.05), and annotated several genes of interest based on their P value and fold change (Fig. 5). We found that a few annotated genes are linked to integrin or CD44 expression/activity. TSPAN1 has relevance toward cancer cell migration, since it is a cancer-associated protein involved in the signaling pathway for survival and invasion.37 Additionally, there was a 16-fold increase in HAS2 in CD44KO cells. Hyaluronan synthase 2 (HAS2), is responsible for producing CD44's extracellular matrix ligand hyaluronic acid, increased 16-fold upon CD44KO. TNFAIP6 is associated with the family of proteins which bind to hyaluronic acid and plays a role in facilitating CD44-dependent invasion in colorectal cancer.38 EDNRB promotes the function of focal adhesion kinase, paxillin, RhoA, and the PI3K pathway.39 Furthermore, EDNRB is linked to tenascin-C, a molecule which is known to block integrin α5-β1.39 LTBP2 interacts with integrin α3β1 solely on fibronectin, as an anti-adhesive.40 Although we do not see direct evidence in altered expression levels for motors or clutches, we see several genes involved in processes and pathways that involve motor and clutch levels/activities. Gene ontology (GO) was also used to observe the 270 significant genes' involvement in biological processes, cellular components, and molecular functions. GO cellular component shows an increase in over 100-fold for genes associated with multiple extracellular and collagen-involved components (supplementary material Table 2).
FIG. 5.
CD44KO fold enrichment compared to WT shows significant differences in gene expression. The smaller graph shows the full volcano plot (left), the gray area contains statistically insignificant data. Here, we zoom into the significant genes that show fold changes with a P < 0.05 and a fold change > 2 (right). Red and blue indicate an increase and decrease, respectively, in gene expression in CD44KO compared to WT.
DISCUSSION
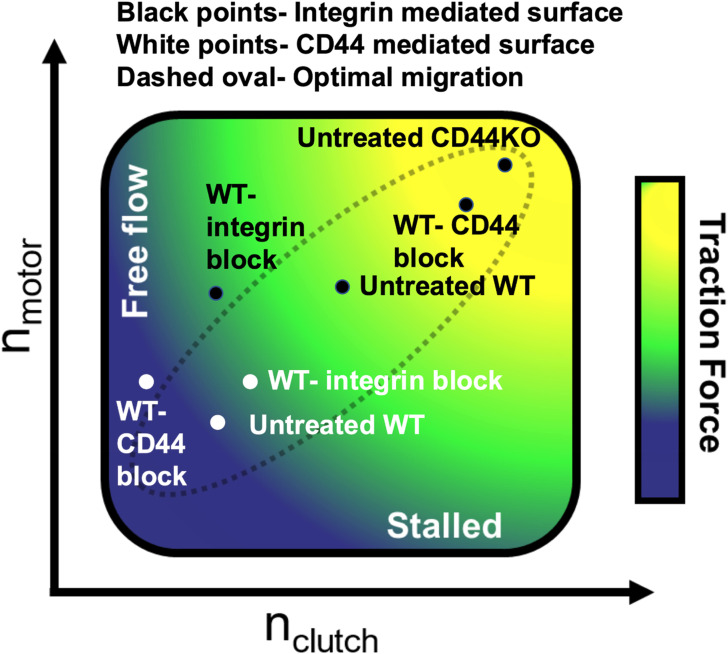
Our data show that when CD44 is unavailable from either inhibition or knockout on collagen I, cell traction significantly increases. The same is observed in CD44-mediated adhesion conditions when integrins are blocked, there is an increase in traction force. Interestingly, we did not observe major changes in migration between WT and CD44KO across the stiffnesses used in the migration assays; however, we see ample cell migration in both cell types. We observe that CD44-integrin crosstalk is impactful on both CD44-mediated and integrin-mediated surfaces in terms of generating traction force, indicating that the two clutch components act in a mutually antagonistic relationship. In the CMS model, the condition with increased motors and clutches matches our experimental results well. We see a substantial increase in cell traction, and a minimal change in migration, along with modest changes to cell shape, all of which are consistent with experimental observations. Our experimental data and the model both support the idea of crosstalk between integrins and CD44, where they maintain an antagonistic relationship between one another. Once one of the receptors is blocked or knocked out, the increase in traction force suggests there is effectively an increase in clutches and motors (Fig. 6). Our model's migration assays support the idea that both motors and clutches are balanced, and both increase in CD44KO on integrin-mediated surfaces (Fig. 6). This allows for a balanced motor-clutch system to be maintained so that there is similar migration between WT and CD44KO. Figure 6 highlights the several conditions observed and shows that migration can occur efficiently at any traction if motors and clutches are balanced.
FIG. 6.
Combination of TFM and migration suggests that CD44-integrin crosstalk increases both clutches and motors. The seven conditions that were tested in TFM are plotted in position based on both the TFM and migration assays. The traction gradient on the schematic was based on our previous modeling.4
Our results seem counterintuitive since CD44 does not adhere to collagen. We expected U251 CD44KO to have either no phenotype or possibly a slight decrease in traction force on integrin-mediated PAGs, since there have been previous reports of direct interactions between collagen and CD44.41,42
If CD44 interacted with collagen in an adhesive role, there would have been a decrease in traction force upon knockout, which was not observed experimentally. At first glance, the data from Fig. 1 is unexpected based on comparisons to a previous study, where adhesion was observed in CD44 knock down U373-MG cells.43 In their study, adhesion strength was determined by the cell's ability to remain adhered to the substrate when centrifugal force was applied to remove it, while we observed traction strain energy, which measured the amount of force exerted to the substrate by the cell. Furthermore, if motors and clutches are changing coordinately like our prediction, adhesion strength is not as affected. U251 cells had CD44 knocked out, while their U373-MG cells only had CD44 knocked down. This is the first study to examine crosstalk in terms of functional effects on force transmission, so the results between the two studies could be unrelated. In another recent study, a novel technique called Rupture and Deliver tension gauge tethers (RAD-TGTs) was used to measure force generation in U251 WT and CD44KO cells.44 They show a slight decrease in RAD-TGT signal for CD44KO compared to WT, on 12pN TGTs and exhibit no change on 54 pN TGTs. RAD-TGTs measure single molecule forces, and the force per bond would not change much if both motors and clutches are increasing. TFM, however, shows the entire cell's forces. According to our hypothesis, we would expect no change in the RAD-TGT experiment, but the slight imbalance in motors and clutches could lead to less rupturing and lower signal. We use a collagen-coated PAG with a Young's modulus of 4.6 kPa for our integrin-mediated traction force assays we, while Kim et al. used a HA hydrogel conjugated with RGD peptide with a shear modulus of 4.6 kPa.43 Pawlak et al. used a glass bottom coated with RAD-TGTs containing WDV fused with echistatin in the other.44 Our data show consistency under both integrin-mediated and CD44-mediated conditions, along with support from our established models and mRNA sequencing, supporting our hypothesis.
By using IM7 to reproduce a CD44-mediated environment, we were able to specifically observe CD44's adherence in response to blocking integrins. However, IM7 is not representative of HA, CD44's natural ECM protein. HA was not technically feasible using our sulfo-SANPAH method, but HA-coated PAGs and HA gels will be further investigated for future work. Furthermore, in one of our assays, we chose to inhibit αV and α3 integrins because they were the most abundant α-integrins in our cell line, according to previous studies.27 According to previous studies,8 neither of these α-integrins bind to collagen. Interestingly when those α-integrins were inhibited, we still saw an effect on collagen.
CONCLUSION
We hypothesized that integrins and CD44 play a role in traction and migration even when they are not directly involved in the adhesion to a surface. In this study, we interrogated CD44-integrin crosstalk and observed a strong mutually antagonistic relationship between the two with respect to traction force but not migration. In several studies, CD44 was suggested to be a target for therapeutic treatments in glioblastoma. The outcomes of the research provided a new insight into the relationship between integrins and CD44, pushing toward multi-targeting therapies that could take advantage of their antagonistic relationship in the context of cell migration. For example, perhaps we can target CD44 to allow integrins to overcompensate for the CD44 loss while also targeting non-muscle myosin II to prevent the motors from coordinating with the clutches. This approach could be used on other cancers characterized by the overexpression of CD44, such as some breast cancers. Future research should include investigating the mechanism for the crosstalk and characterizing its signaling pathway.
METHODS
U251 glioma cell culture
U251 glioma cells were cultured in vented T75 flasks in a humidified incubator under conditions of 37 °C, 5% CO2. U251 media was Dulbecco's modified Eagle's medium (Gibco 11-320-033) supplemented with 9% heat-inactivated fetal bovine serum (Gibco A3840001) and 1% penicillin/streptomycin solution (Cellgro 30-001-CI). To prevent over-confluency, cells were treated with 0.25% trypsin with EDTA in Hanks' balanced salt solution. Cells were then spun down in a centrifuge at 220×g for 5 min, decanted, and resuspended in 5 ml of fresh media and placed back in a flask. During cell seeding, cells were plated at ∼10 000 cells per gel onto UV sterilized PAGs. Then, they were incubated for at least 2 h before imaging to allow for adequate cell adhesion. U251 CD44KO cells were obtained via CRISPR as previously described.44 CD44KO was obtained via CRISPR as previously described.
RNA sequencing
Cells were cultured under standard U251 glioma conditions as mentioned above until ∼1 × 106 cells in total. Cells were flash frozen and sent to the University of Minnesota Genomic Center. RNA was extracted using QIAGEN RNeasy plus universal kit. Then, purity and concentration was assessed using UV spectroscopy (NanoDrop) and fluorimetry (PicoGreen quantification). RNA integrity was also determined using the Agilent 2100 Bioanalyzer. They used Illumina TruSeq mRNA kit to sequence RNA.
Preparing polyacrylamide gels
No. 0 glass bottom culture dishes (MatTek P35G-0-20-C) were treated with 97% (3-aminoproyl) trimethoxysilane (Aldrich 281778), and 0.5% glutaraldehyde (Polysciences 01909) to activate the glass, causing PAGs to be able to adhere firmly to the glass bottom. Once silanized, dishes can be stored for up to 2 months in a desiccator.
Polyacrylamide gels were first mixed into a prepolymer using the five following ingredients: 40% acrylamide solution (Fisher BP1402), 2% bis-acrylamide solution (Fisher BP1404), 1 M 4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES, Sigma H6147) solution, sonicated or vigorously mixed 200 nm fluorosphere crimson beads solution (Invitrogen F-8810), and de-ionized water. In summary, 4.6 kPa PAGs contained 10% acrylamide solution, 10% bis-acrylamide solution, 1% HEPES, 1% fluorosphere crimson beads, 0.6% APS, 0.4% TEMED, and 78% diH2O. For 9.3 kPa PAGs, 12.5% acrylamide solution, 5% bis-acrylamide solution, 1% HEPES, 1% fluorosphere crimson beads, 0.6% APS, 0.4% TEMED, and 80% diH2O were present. Finally, 20 kPa PAGs contained 25% acrylamide solution, 5% bis-acrylamide solution, 1% HEPES, 1% fluorosphere crimson beads, 0.6% APS, 0.4% TEMED, and 68% diH2O. For migration assays, additional diH2O was used in the place 200 nm crimson fluorospheres. We adjusted the ratio of acrylamide and bis-acrylamide to create gels with varying Young's modulus.
Next, we degassed the solution for 30 min using a desiccator to aid in the polymerization process. During the polymerization step, the solution is removed from the desiccator and 1% ammonium persulfate (Bio-Rad 161-0700) solution and N,N,N′,N′-tetramethylethylenediamine (TEMED, Fisher BP150) are added to the solution. Once added, the polymerization will initiate, so we then quickly pipet 5 μl onto a silanized glass bottom dish and cover it with a glass coverslip that was cleaned with 70% ethanol. For TFM assays, the solutions are flipped over during polymerization so that the fluorospheres could be at the top of the gel once polymerization is complete. Once gels formed, 50 mM HEPES was diluted in DI water to wash the gels and stored at 4 °C overnight.
The second step in gel preparation was coating the gels with the protein of interest. In this study, we use collagen I (CB40236) or a CD44Ab IM7 (BDB553131) as adhesive coatings, for integrin-mediated or CD44-mediated conditions, respectively. The next day, PAGs were treated with 0.5 mg/ml sulfosuccinimidyl 6–(4′-azido-2′-nitrophenylamino) hexanoate (sulfo-SANPAH, Thermo 22589) to functionalize the surface and allow the protein of interest to undergo covalent bonding to the PAG surface. After treatment, the adhesive protein of interest is added to PAG and left at 4 °C overnight. In the morning of cell seeding, the PAG is washed off with PBS and sterilized using UV light for ∼15 min. Finally, fresh media and cells are added to the newly functionalized PAG and placed in the incubator. For collagen type I coating, a 0.2 mg/ml solution, diluted with PBS (pH 7.4) was used to serve as an integrin-mediated ECM. For CD44ab coating, a 0.02 mg/ml solution of IM7 was diluted with PBS as described above.
Traction force microscopy
PAGs were made on a Young's modulus of 4.6 kPa, with crimson fluorospheres added to the PAG solution. After seeding, cells were incubated for at least 3 h before imaging. Cells were imaged using a Nikon Eclipse Ti2 inverted microscope and CoolSnap HQ2 CCD camera (Photometrics). A Bold Line top humidified stage incubator (Okolab) was used at 37 C° and 5% CO2. For traction force microscopy, a Plan Apo 40×/0.95 NA objective with a 1.5× magnification lens was used to image individual cells. Briefly, phase-contrast images of cells, along with mCherry filter fluorescence images of the crimson fluorospheres before and after cell adhesion is disrupted by 15-min trypsinization treatment. Once images were acquired, a custom MATLAB code was used for the traction force analysis as previously described.27 Briefly, images are aligned, cropped, and bead displacement is measured via cross correlation analysis to calculate traction strain energy using Fourier transform traction cytometry.
Migration assays
PAGs were made on a wide range of stiffnesses, from 0.7, 4.6, 20, 100, to 200 kPa. Like above, after seeding, the cells were incubated for at least 3 h. For migration assays, a Plan Fluor 10×/0.30 NA objective with phase contrast was used to create time-lapse movies. Images were taken on five fields per dish at 15-min intervals for 16 h. To quantify the migration of this random walk, we use the mean squared displacement to measure the displacement of a cell over time. We then calculate the random motility coefficient from the slope of the trendline for MSD over time.
Simulations
The cell migration simulator, which is based on the motor-clutch model for cell motility, was used as previously described.27 Parameters chosen for the three cases, WT, WT + enhanced clutches, and WT + enhanced motors and clutches, can be found in the Supplementary information.
Statistical analysis
Analysis of all experimental traction force microscopy assays tested statistical differences by using the Mann–Whitney test. Red lines indicate the median. Migration assays were tested using the Kruskal–Wallis test. The middle red line indicates the median, and the top and bottom lines represent the 25th and 75th percentiles, respectively. Computational data are analyzed using the mean and s.e.m.
SUPPLEMENTARY MATERIAL
See the supplementary material for details on two supplementary tables and one supplementary figure.
ACKNOWLEDGMENTS
Research reported in this publication was supported by Grant Nos. U54CA210190, P01CA254849, and U54CA268069. The content of this work is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Note: This paper is part of the special issue on Physical Sciences Approaches to Cancer Research.
Contributor Information
Wendy R. Gordon, Email: mailto:wrgordon@umn.edu.
David J. Odde, Email: mailto:oddex002@umn.edu.
AUTHOR DECLARATIONS
Conflict of Interest
The authors have no conflicts to disclose.
Ethics Approval
Ethics approval is not required.
Author Contributions
Marcus D. Kelly: Data curation (equal); Formal analysis (equal); Methodology (equal); Project administration (equal); Writing – original draft (equal). Matthew R. Pawlak: Data curation (supporting). Kevin H. Zhan: Data curation (supporting); Writing – review & editing (supporting). Ghaidan A. Shamsan: Data curation (supporting). Wendy R. Gordon: Conceptualization (equal); Supervision (equal); Writing – review & editing (equal). David J. Odde: Conceptualization (equal); Supervision (equal); Writing – review & editing (equal).
DATA AVAILABILITY
The data that support the findings of this study are available from the corresponding authors upon reasonable request.
References
- 1. Hanahan D., “ Hallmarks of cancer: New dimensions,” Cancer Discovery 12(1), 31–46 (2022). 10.1158/2159-8290.CD-21-1059 [DOI] [PubMed] [Google Scholar]
- 2. Chen N., Cheng P., and Dan L., “ Epigenetic underpinnings of inflammation: A key to unlock the tumor microenvironment in glioblastoma,” Front. Immunol. 13, 869307 (2022). 10.3389/fimmu.2022.869307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vicente-Manzanares M., Ma X., Adelstein R. S., and Horwitz A. R., “ Non-muscle myosin II takes centre stage in cell adhesion and migration.” Nat. Rev. Mol. Cell Biol. 10(11), 778–790 (2009). 10.1038/nrm2786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bangasser B. L., Rosenfeld S. S., and Odde D. J., “ Determinants of maximal force transmission in a motor-clutch model of cell traction in a compliant microenvironment,” Biophys. J. 105(3), 581–592 (2013). 10.1016/j.bpj.2013.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mitchison T. and Kirschner M., “ Cytoskeletal dynamics and nerve growth,” Neuron 1(9), 761–772 (1988). 10.1016/0896-6273(88)90124-9 [DOI] [PubMed] [Google Scholar]
- 6. Liu Y. J., Le Berre M., Lautenschlaeger F., Maiuri P., Callan-Jones A., Heuzé M., Takaki T., Voituriez R., and Piel M., “ Confinement and low adhesion induce fast amoeboid migration of slow mesenchymal cells,” Cell 160(4), 659–672 (2015). 10.1016/j.cell.2015.01.007 [DOI] [PubMed] [Google Scholar]
- 7. Ridley A. J., Schwartz M. A., Burridge K., Firtel R. A., Ginsberg M. H., Borisy G., Parsons T. J., and Horwitz A. R., “ Cell migration: Integrating signals from front to back,” Science 302(5651), 1704–1709 (2003). 10.1126/science.1092053 [DOI] [PubMed] [Google Scholar]
- 8. Hynes R. O., “ Integrins: Bidirectional, allosteric signaling machines,” Cell 110(6), 673–687 (2002). 10.1016/S0092-8674(02)00971-6 [DOI] [PubMed] [Google Scholar]
- 9. Gallant N. D., Michael K. E., and García A. J., “ Cell adhesion strengthening: Contributions of adhesive area, integrin binding, and focal adhesion assembly,” Mol. Biol. Cell 16(9), 4329–4340 (2005). 10.1091/mbc.e05-02-0170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chinthalapudi K., Rangarajan E. S., and Izard T., “ The interaction of talin with the cell membrane is essential for integrin activation and focal adhesion formation,” Proc. Nat. Acad. Sci. U. S. A. 115(41), 10339–10344 (2018). 10.1073/pnas.1806275115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wegener K. L., Partridge A. W., Han J., Pickford A. R., Liddington R. C., Ginsberg M. H., and Campbell I. D., “ Structural basis of integrin activation by talin,” Cell 128(1), 171–182 (2007). 10.1016/j.cell.2006.10.048 [DOI] [PubMed] [Google Scholar]
- 12. Ziegler W. H., Gingras A. R., Critchley D. R., and Emsley J., “ Integrin connections to the cytoskeleton through talin and vinculin,” Biochem. Soc. Trans. 36(2), 235–239 (2008). 10.1042/BST0360235 [DOI] [PubMed] [Google Scholar]
- 13. Kim D. H. and Wirtz D., “ Focal adhesion size uniquely predicts cell migration,” FASEB J. 27(4), 1351–1361 (2013). 10.1096/fj.12-220160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Miranti C. K., Leng L., Maschberger P., Brugge J. S., and Shattil S. J., “ Identification of a novel integrin signaling pathway involving the kinase Syk and the guanine nucleotide exchange factor Vav1,” Curr. Biol. 8(24), 1289–1299 (1998). 10.1016/S0960-9822(07)00559-3 [DOI] [PubMed] [Google Scholar]
- 15. Geiger B., Bershadsky A., Pankov R., and Yamada K. M., “ Transmembrane crosstalk between the extracellular matrix and the cytoskeleton,” Nat. Rev. Mol. Cell Biol. 2(11), 793–805 (2001). 10.1038/35099066 [DOI] [PubMed] [Google Scholar]
- 16. Critchley D. R., “ Cytoskeletal proteins talin and vinculin in integrin-mediated adhesion,” Biochem. Soc. Trans. 32(Pt 5), 831–836 (2004). 10.1042/BST0320831 [DOI] [PubMed] [Google Scholar]
- 17. Boujemaa-Paterski R., Martins B., Eibauer M., Beales C. T., Geiger B., and Medalia O., “ Talin-activated vinculin interacts with branched actin networks to initiate bundles,” ELife 9, e53990 (2020). 10.7554/eLife.53990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Choi S. H., Takahashi K., Eto H., Yoon S. S., and Tanabe K. K., “ CD44s expression in human colon carcinomas influences growth of liver metastases,” Int. J. Cancer 85(4), 523–526 (2000). [DOI] [PubMed] [Google Scholar]
- 19. Zeilstra J., Joosten S. P. J., van Andel H., Tolg C., Berns A., Snoek M., van de Wetering M., Spaargaren M., Clevers H., and Pals S. T., “ Stem cell CD44v isoforms promote intestinal cancer formation in Apc(Min) mice downstream of Wnt signaling,” Oncogene 33(5), 665–670 (2014). 10.1038/onc.2012.611 [DOI] [PubMed] [Google Scholar]
- 20. Louderbough J. M. V. and Schroeder J. A., “ Understanding the dual nature of CD44 in breast cancer progression,” Mol. Cancer Res. 9(12), 1573–1586 (2011). 10.1158/1541-7786.MCR-11-0156 [DOI] [PubMed] [Google Scholar]
- 21. Sneath R. J. and Mangham D. C., “ The normal structure and function of CD44 and its role in neoplasia,” Mol. Pathol. 51(4), 191–200 (1998). 10.1136/mp.51.4.191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ishii S., Ford R., Thomas P., Nachman A., Steele G., and Jessup J. M., “ CD44 participates in the adhesion of human colorectal carcinoma cells to laminin and type IV collagen,” Surg. Oncol. 2(4), 255–264 (1993). 10.1016/0960-7404(93)90015-Q [DOI] [PubMed] [Google Scholar]
- 23. Lv L., Liu H. G., Dong S. Y., Yang F., Wang Q. X., Guo G. L., Pan Y. F., and Zhang X. H., “ Upregulation of CD44v6 contributes to acquired chemoresistance via the modulation of autophagy in colon cancer SW480 cells,” Tumour Biol. 37(7), 8811–8824 (2016). 10.1007/s13277-015-4755-6 [DOI] [PubMed] [Google Scholar]
- 24. Thorne R. F., Legg J. W., and Isacke C. M., “ The role of the CD44 transmembrane and cytoplasmic domains in co-ordinating adhesive and signalling events,” J. Cell Sci. 117(3), 373–380 (2004). 10.1242/jcs.00954 [DOI] [PubMed] [Google Scholar]
- 25. Palecek S. P., Loftus J. C., Ginsberg M. H., Lauffenburger D. A., and Horwitz A. F., “ Integrin-ligand binding properties govern cell migration speed through cell-substratum adhesiveness,” Nature 385(6616), 537–540 (1997). 10.1038/385537a0 [DOI] [PubMed] [Google Scholar]
- 26. DiMilla P. A., Barbee K., and Lauffenburger D. A., “ Mathematical model for the effects of adhesion and mechanics on cell migration speed,” Biophys. J. 60(1), 15–37 (1991). 10.1016/S0006-3495(91)82027-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bangasser B. L., Shamsan G. A., Chan C. E., Opoku K. N., Tüzel E., Schlichtmann B. W., Kasim J. A. et al. , “ Shifting the optimal stiffness for cell migration,” Nat. Commun. 8, 15313 (2017). 10.1038/ncomms15313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Haeger A., Wolf K., Zegers M. M., and Friedl P., “ Collective cell migration: Guidance principles and hierarchies,” Trends Cell Biol. 25(9), 556–566 (2015). 10.1016/j.tcb.2015.06.003 [DOI] [PubMed] [Google Scholar]
- 29. Ulrich T. A., Juan Pardo E. M., and Kumar S., “ The mechanical rigidity of the extracellular matrix regulates the structure, motility, and proliferation of glioma cells,” Cancer Res. 69(10), 4167–4174 (2009). 10.1158/0008-5472.CAN-08-4859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Peyton S. R. and Putnam A. J., “ Extracellular matrix rigidity governs smooth muscle cell motility in a biphasic fashion,” J. Cell. Physiol. 204(1), 198–209 (2005). 10.1002/jcp.20274 [DOI] [PubMed] [Google Scholar]
- 31. Lacouture M. E., Schaffer J. L., and Klickstein L. B., “ A comparison of type I collagen, fibronectin, and vitronectin in supporting adhesion of mechanically strained osteoblasts,” J. Bone Miner. Res. 17(3), 481–492 (2002). 10.1359/jbmr.2002.17.3.481 [DOI] [PubMed] [Google Scholar]
- 32. Culty M., “ The hyaluronate receptor is a member of the CD44 (H-CAM) family of cell surface glycoproteins [published Erratum appears in J. Cell Biol. 1991 Feb;112(3): Following 513],” J. Cell Biol. 111(6), 2765–2774 (1990). 10.1083/jcb.111.6.2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hutás G., Bajnok É., Gál I., Finnegan A., Glant T. T., and Mikecz K., “ CD44-specific antibody treatment and CD44 deficiency exert distinct effects on leukocyte recruitment in experimental arthritis,” Blood 112(13), 4999–5006 (2008). 10.1182/blood-2008-04-150383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fujisaki T., Tanaka Y., Fujii K., Mine S., Saito K., Yamada S., Yamashita U., Irimura T., and Eto S., “ CD44 stimulation induces integrin-mediated adhesion of colon cancer cell lines to endothelial cells by up-regulation of integrins and c-Met and activation of integrins,” Cancer Res. 59(17), 4427–4434 (1999). [PubMed] [Google Scholar]
- 35. Lekka M., Gnanachandran K., Kubiak A., Zieliński T., and Zemła J., “ Traction force microscopy—Measuring the forces exerted by cells,” Micron 150, 103138 (2021). 10.1016/j.micron.2021.103138 [DOI] [PubMed] [Google Scholar]
- 36. Butler J. P., Tolić-Nørrelykke I. M., Fabry B., and Fredberg J. J., “ Traction fields, moments, and strain energy that cells exert on their surroundings,” Am. J. Physiol. 282(3), C595–C605 (2002). 10.1152/ajpcell.00270.2001 [DOI] [PubMed] [Google Scholar]
- 37. Hou F. Q., Lei X. F., Yao J. L., Wang Y. J., and Zhang W., “ Tetraspanin 1 is involved in survival, proliferation and carcinogenesis of pancreatic cancer,” Oncol. Rep. 34(6), 3068–3076 (2015). 10.3892/or.2015.4272 [DOI] [PubMed] [Google Scholar]
- 38. Chen H., Wu Y., Jiang Y., Chen Z., and Zheng T., “ DKC1 aggravates gastric cancer cell Migration and invasion through up-regulating the expression of TNFAIP6,” Funct. Integr. Genomics 24(2), 38 (2024). 10.1007/s10142-024-01313-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lange K., Kammerer M., Hegi M. E., Grotegut S., Dittmann A., Huang W., Fluri E. et al. , “ Endothelin receptor type B counteracts tenascin-C-induced endothelin receptor type A-dependent focal adhesion and actin stress fiber disorganization,” Cancer Res. 67(13), 6163–6173 (2007). 10.1158/0008-5472.CAN-06-3348 [DOI] [PubMed] [Google Scholar]
- 40. Hyytiäinen M. and Keski-Oja J., “ Latent TGF-β binding protein LTBP-2 decreases fibroblast adhesion to fibronectin,” J. Cell Biol. 163(6), 1363–1374 (2003). 10.1083/jcb.200309105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Goodison S., Urquidi V., and Tarin D., “ CD44 cell adhesion molecules,” Mol. Pathol. 52(4), 189–196 (1999). 10.1136/mp.52.4.189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lauer-Fields J. L., Malkar N. B., Richet G., Drauz K., and Fields G. B., “ Melanoma cell CD44 interaction with the Α1(IV)1263–1277 region from basement membrane collagen is modulated by ligand glycosylation,” J. Biol. Chem. 278(16), 14321–14330 (2003). 10.1074/jbc.M212246200 [DOI] [PubMed] [Google Scholar]
- 43. Kim Y. and Kumar S., “ CD44-mediated adhesion to hyaluronic acid contributes to mechanosensing and invasive motility,” Mol. Cancer Res. 12(10), 1416–1429 (2014). 10.1158/1541-7786.MCR-13-0629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pawlak M. R., Smiley A. T., Ramirez M. P., Kelly M. D., Shamsan G. A., Anderson S. M., Smeester B. A., Largaespada D. A., Odde D. J., and Gordon W. R., “ RAD-TGTs: High-throughput measurement of cellular mechanotype via rupture and delivery of DNA tension probes,” Nat. Commun. 14(1), 2468 (2023). 10.1038/s41467-023-38157-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding authors upon reasonable request.