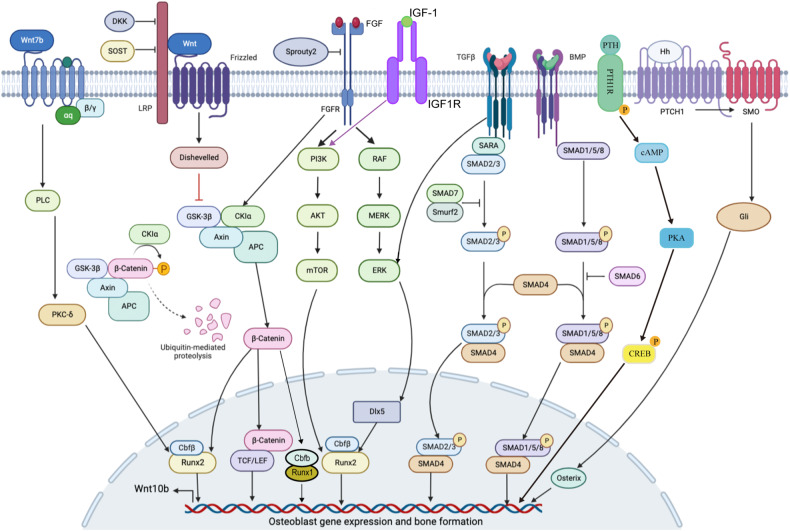
Fig. 2. Canonical signaling pathways in osteoblast differentiation.
Several canonical signaling pathways control the activity of key transcription factors to mediate osteoblast differentiation. Wnt, TGF-β, BMP, FGF, and Hedgehog pathways are the most classic pathways that have been studied during osteoblast differentiation. Wnt binds with FZD receptors, causing the β-catenin accumulation. β-catenin then moves to the nucleus, in which it causes target genes to be transcribed. Wnt signaling also regulates Runx1 and Runx2 functions. TGF-β and BMP signaling regulate osteoblast-specific gene expression through multiple Smad proteins. TGF-β signaling mainly activates Smad2/3, while BMP signaling activates Smad1/5/8. FGF and FGFR can also regulate osteoblast differentiation and osteoblast-specific gene expression via downstream pathways such as PI3K-AKT and ERK pathways. Runx2, Osterix, and several other transcription factors are also necessary for osteoblast differentiation, and these transcription factors are regulated by these classic pathways. Hedgehog signaling is activated through Hh ligand binding to the 12-transmembrane receptor Patched 1 (PTCH1), which relieves inhibition of the seven-pass transmembrane G protein-coupled receptor Smoothened (SMO). Actived SMO can initiate the intracellular cascade leading to the activation of three Gli transcription factors, and Gli can then translocate into the nucleus and regulate Osx activation, thereby modulating osteoblast-specific gene expression. PTH regulates osteoblast differentiation by binding with PTH1R, subsequently activating cAMP and PKA, leading to the phosphorylation of CREB to regulate osteoblast-specific gene expression. IGF-1 binds to its receptor IGF1R and activates PI3K-Akt pathway, resulting in the activation of mTOR and promotion of osteoblast differentiation.