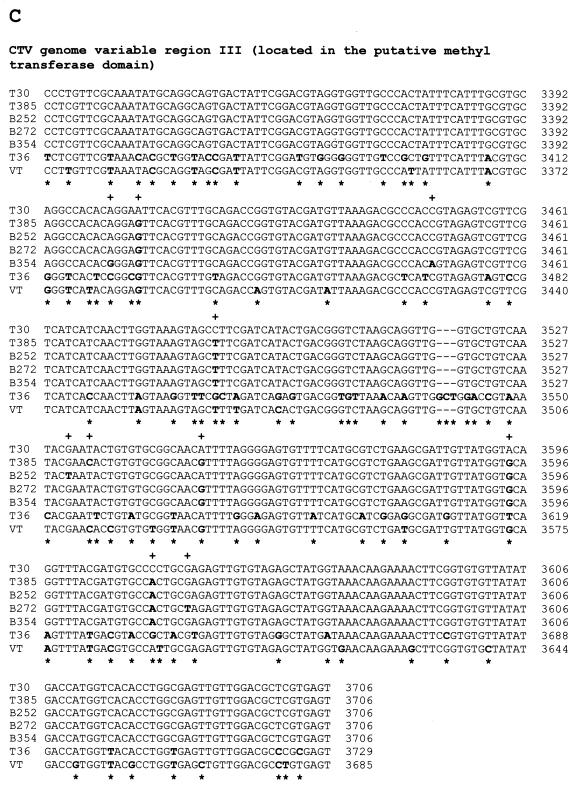
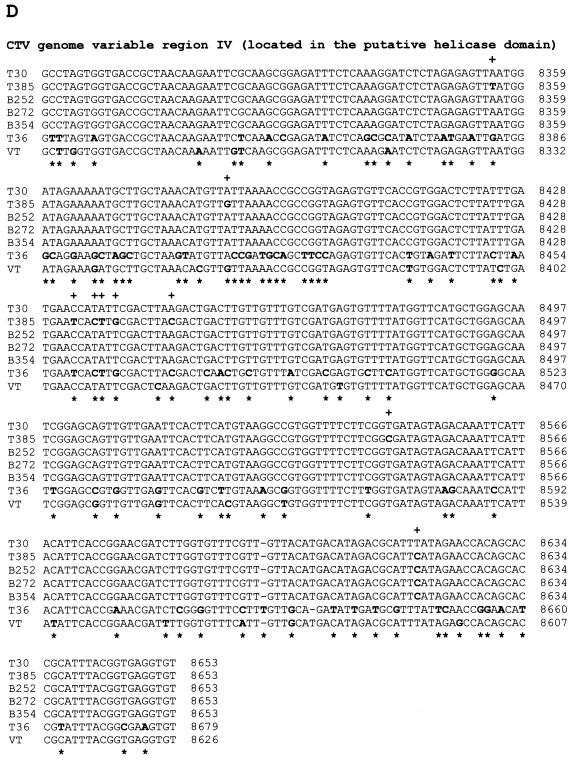
FIG. 2.
Nucleotide sequence alignments of CTV hypervariable genomic regions (ClustalW program alignments; see Materials and Methods). (A) Alignment of variable region I (5′ NTR, 108 nt) sequences. (B) Alignment of variable region II (located from nt 109 to nt 324 of the T30 genome in the 5′-proximal region of the replicase gene) sequences. (C) Alignment of variable region III (located from nt 3324 to nt 3706 of the T30 genome in the putative methyltransferase domain) sequences. (D) Alignment of variable region IV (located from nt 8265 to nt 8627 of the T30 genome in the putative helicase domain of the replicase ORF) sequences. In all panels, nucleotide differences in aligned sequences are showed in boldface. +, nucleotide differences between T30 and the mild CTV isolates T385, B252, B272, and B354; ∗, nucleotide differences between T30 and severe isolates T36 and VT.