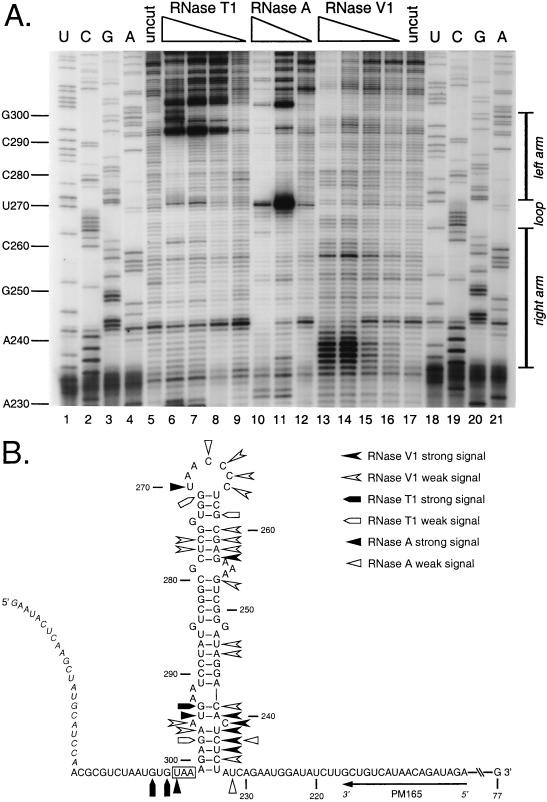
FIG. 5.
Enzymatic structural probing of the bulged stem-loop structure. A 264-nt RNA encompassing the stem-loop region of the MHV 3′ UTR was transcribed in vitro from plasmid pBL122 (Fig. 2A), purified, renatured, and digested with various concentrations of RNases. Positions of cleavage sites were determined by primer extension with a 5′-end-labeled primer, PM165, complementary to nt 199 to 216. (A) Enzymatic cleavage sites generated by single-stranded nucleases RNase T1 (G specific) and RNase A (U and C specific) and double-stranded nuclease RNase V1. Lanes 1 to 4 and 18 to 21, sequencing ladders generated with end-labeled PM165 and terminated with ddATP, ddGTP, ddCTP, or ddTTP, respectively; lanes 5 and 17, undigested RNA; lanes 6 to 9, RNA digested with 15, 10, 5.0, and 1.0 U of RNase T1, respectively; lanes 10 to 12, RNA digested with 0.01, 0.001, and 0.0001 U of RNase A, respectively; lanes 13 to 16, RNA digested with 0.5, 0.3, 0.1, and 0.05 U of RNase V1, respectively. Ten-nucleotide intervals and the position of the stem-loop are indicated to the left and right of the autoradiogram, respectively. Each primer extension product from nuclease-digested RNA terminates one base downstream of the corresponding nucleotide in the sequencing ladder because all three RNases cut 3′ to their target bases. (B) Summary of observed enzymatic cleavage sites superimposed on the originally proposed stem-loop structure. Nucleotide numbering begins at the 3′ end of the genome, excluding the poly(A) tail; the N gene stop codon is boxed, and the position of the primer PM165 is shown. Bases indicated in italics at the 5′ end of the synthetic RNA are those derived from the polylinker of the transcription vector. Artifactual RNase T1 signals at A293 and U295 are not included in this diagram.