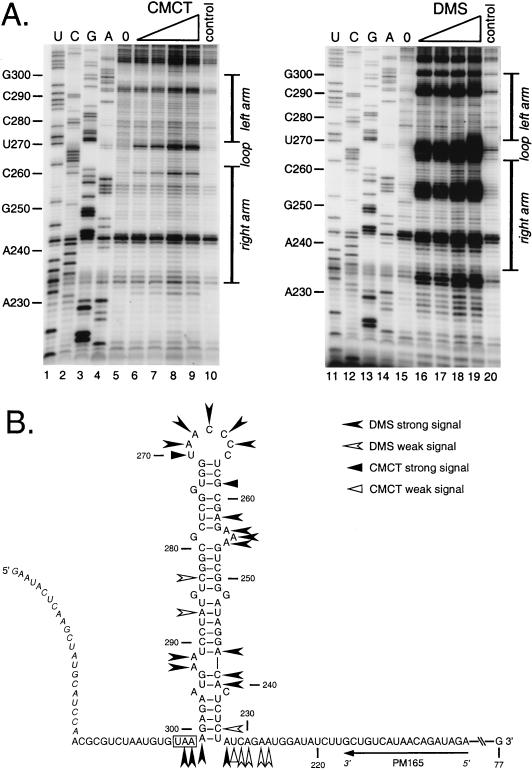
FIG. 6.
Chemical structural probing of the bulged stem-loop structure. A 264-nt RNA encompassing the stem-loop region of the MHV 3′ UTR was transcribed in vitro from plasmid pBL122 (Fig. 2A), purified, renatured, and reacted with various concentrations of chemical reagents. Positions of base modification were determined by primer extension as for Fig. 5. (A) Modification sites generated by single-stranded RNA-specific reagents CMCT (G and U specific) and DMS (A and C specific). Lanes 1 to 4 and 11 to 14, sequencing ladders generated with end-labeled PM165 and terminated with ddATP, ddGTP, ddCTP, or ddTTP, respectively; lanes 5 and 15, unmodified RNA; lanes 6 to 9, RNA modified with 4.2, 8.4, 12.6, and 16.8 mg of CMCT per ml, respectively; lanes 16 to 19, RNA modified with 0.5, 1.0, 1.5, and 2.0% DMS, respectively; lanes 10 and 20, control reactions in which the highest concentration of each reagent was added to RNA after addition of quenching reagents. Ten-nucleotide intervals and the position of the stem-loop are indicated to the left and right of each autoradiogram, respectively. Each primer extension product from chemically modified RNA is positioned one base downstream of the corresponding nucleotide in the sequencing ladder because primer extension terminates at the nucleotide immediately 3′ to the modified base. (B) Summary of observed chemical modification sites superimposed on the originally proposed stem-loop structure. Nucleotide numbering begins at the 3′ end of the genome, excluding the poly(A) tail; the N gene stop codon is boxed, and the position of the primer PM165 is shown. Bases indicated in italics at the 5′ end of the synthetic RNA are those derived from the polylinker of the transcription vector. Artifactual DMS signals at U234, G257, U270, G271, and G294 are not included in this diagram.