Figure 1. Methodological overview.
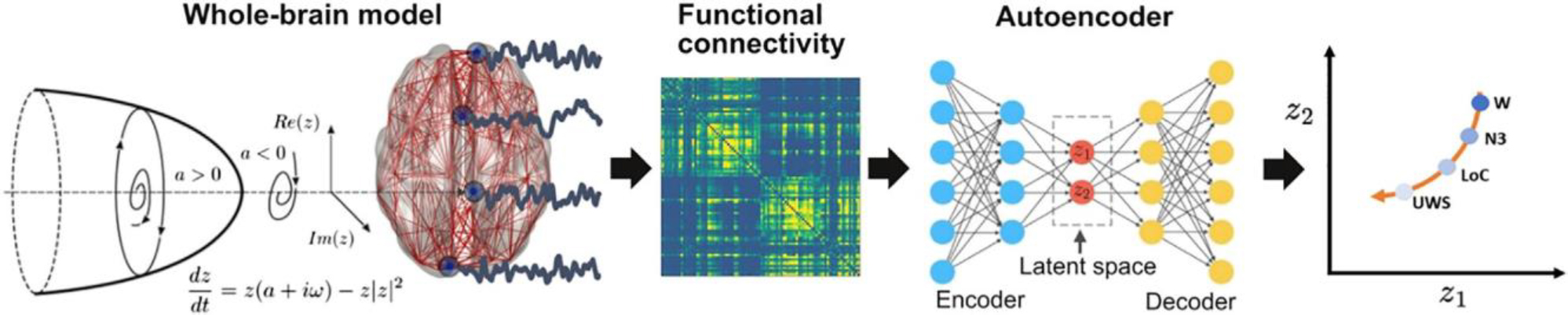
A whole-brain model with local dynamics given by Hopf bifurcations was implemented at nodes defined by the AAL parcellation, coupled with the anatomical connectome. We included spatial heterogeneity based on RSN in the model parameters. The model was tuned to reproduce the empirical FC for each condition, and the resulting parameters were used to generate a surrogate database of simulated FC matrices that were represented in a latent space using a VAE. Finally, perturbations were introduced in the model as an external periodic force, resulting in a set of trajectories in latent space (one per pair of homotopic AAL regions) parameterized accordingly the amplitude of the forcing parameter.
