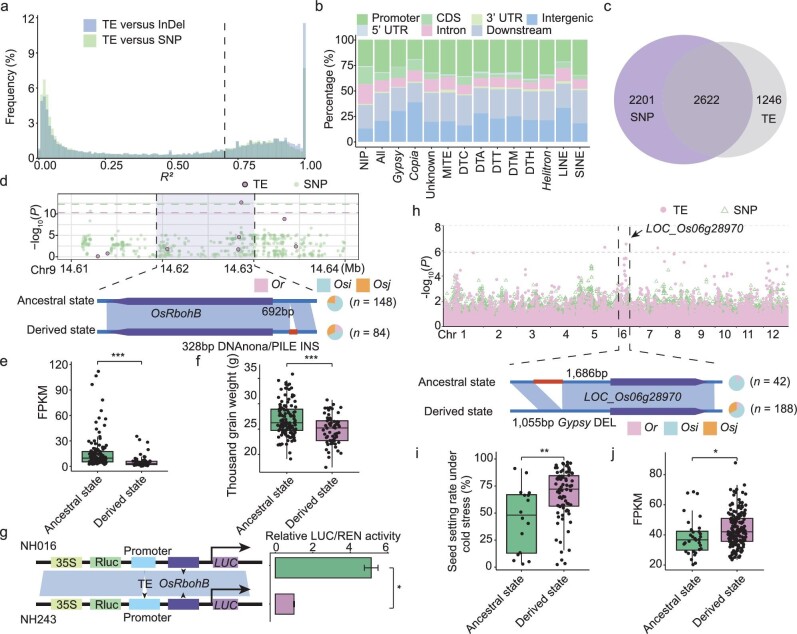
Figure 4.
TE variations associated with gene expression and agricultural traits. (a) The distribution of linkage disequilibrium values (LD, R²) between dTEs and SNPs/InDels within 50 kb of the dTEs. For each dTE, the maximum R² with adjacent SNPs/InDels (within 50 kb on either side) was recorded. The dashed line indicates R² = 0.70. (b) Percentage of dTEs overlapping with genic regions. Genic regions include gene body (CDS, 3’UTR, 5’UTR and intron), and the regions within 2 kb upstream (henceforth ‘promoter’) and 2 kb downstream (henceforth ‘downstream’) of the gene body. Unknown refers to LTR/unknown family. ‘All’ refers to the percentage of dTEs in the genic regions of all dTE loci identified in the present study, and ‘NIP’ refers to the percentage of TEs overlapping with genic regions in the Nipponbare genome. (c) Number of eGenes associated with dTE and SNP variants. eGenes are genes whose expression is significantly associated with dTE and SNP variants. (d) Manhattan plot of OsRbohB expression level and the dTE variants and SNPs (top). The leading dTE (328 bp) insertion (INS) associated with OsRbohB expression level is indicated (bottom). (e–f) Differences in the expression level of OsRbohB (e) and the thousand grain weight (f) between accessions with the ancestral state of the dTE INS event (n = 125 and n = 121, respectively) and those with the derived state (both n = 61). Significance was tested by the Student's t-test, ***P < 0.001. (g) Transcriptional activation assays by contransfecting rice protoplasts. Error bars represent the mean ± SD of three biological replicates. (h) GWAS for seed setting rate under cold stress using the TE and SNP data sets for the Os accessions. A locus on chromosome 6, which was identified by TEs but not by SNPs, was significantly associated with seed setting rate under cold stress. The triangles represent SNPs and dots represent dTEs. The most strongly associated dTE was a 1.0 kb dTE (Gypsy) deletion (DEL) event in the promoter of LOC_Os06g28970. (i and j) Comparison of seed setting rate under cold stress (i) and the expression level of LOC_Os06g28970 (j) between accessions with (i.e. derived state, n = 16 and n = 32, respectively) and without (i.e. ancestral state, n = 83 and n = 169, respectively) the dTE DEL event. Significance was tested by the Student's t-test, **P < 0.01, *P < 0.05.