Figure 2: Extracellular loop 2 of GPR161 occupies classic GPCR orthosteric site.
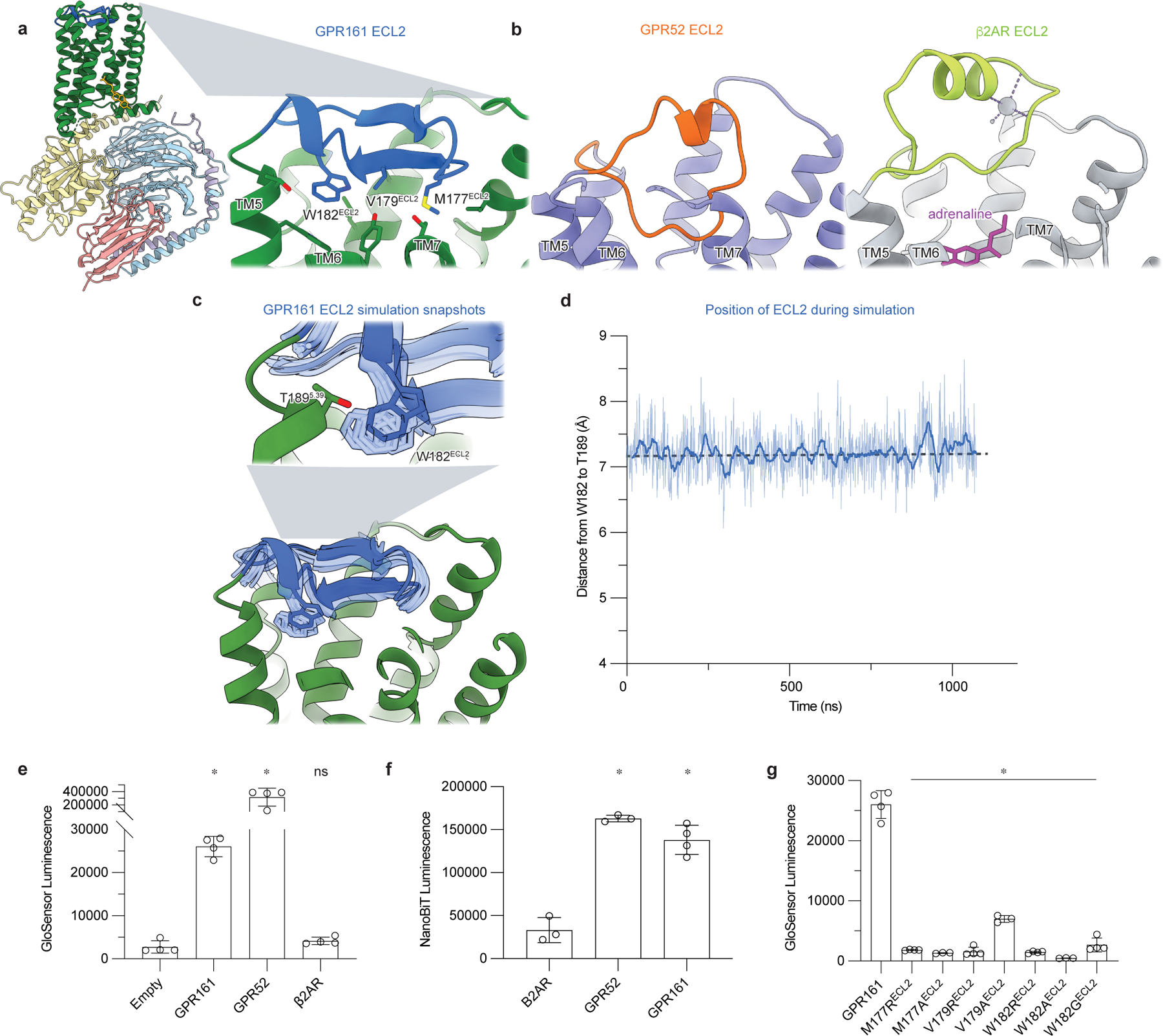
a) The ECL2 of GPR161 makes hydrophobic contacts with the core of the receptor. b) Comparison of ECL2 of the self-activating orphan GPCR GPR52 (PDB ID: 6LI330) and the prototypical agonist-activated GPCR β2-adrenergic receptor (β2AR) bound to agonist adrenaline (PDB ID: 4LDO78). c) In molecular dynamics simulations of GPR161, ECL2 remains in the canonical ligand binding pocket even when mini Gs is removed. Simulation snapshots of ECL2 are shown in light blue, and the cryo-EM structure in dark blue and green. d) Time trace of the distance between residues W182 in ECL2 and T189 in TM4 during a representative molecular dynamics simulation of GPR161 without mini Gs. Thick trace represents a 20 -ns moving average; thin trace represents unsmoothed values. Dashed horizontal line indicates the corresponding distance in the cryo-EM structure. See Methods. Data from remaining simulations is shown in Extended Data Figure 5. e) cAMP production assay for GPR161 (unfused, wildtype), GPR52, and β2AR. GPR161 is constitutively active for cAMP production. Data are mean ± sd, n=3 (for GPR52) or n=4 (for empty, GPR161 and β2AR) biologically independent samples (*P < 0.05; ns, not significant; one-way ANOVA followed by Dunnett’s multiple comparison tests; adjusted P values: GPR161 and GPR52 vs. empty=<0.0001, β2AR vs. empty= 0.9239). f) Nanoluc complementation assay for receptor recruitment of miniGs. Both GPR52 and GPR161 constitutively recruit miniGs. Data are mean ± sd, n=3 (for β2AR, P = <0.0001 and GPR52) or n=4 (for GPR161) biologically independent samples (*P < 0.05; ns, not significant; one-way ANOVA followed by Dunnett’s multiple comparison tests; adjusted P-values: GPR52 and GPR161 vs. β2AR=<0.0001). g) cAMP production assay assessing mutations in ECL2 of GPR161 for residues that make hydrophobic contacts with the transmembrane bundle. Data are mean ± sd, n=4 biologically independent samples (*P < 0.05; ns, not significant; one-way ANOVA followed by Dunnett’s multiple comparison tests; adjusted P values: <0.0001 for all mutants vs. WT GPR161).
