Figure 3: GPR161-miniGs stably and specifically binds cholesterol.
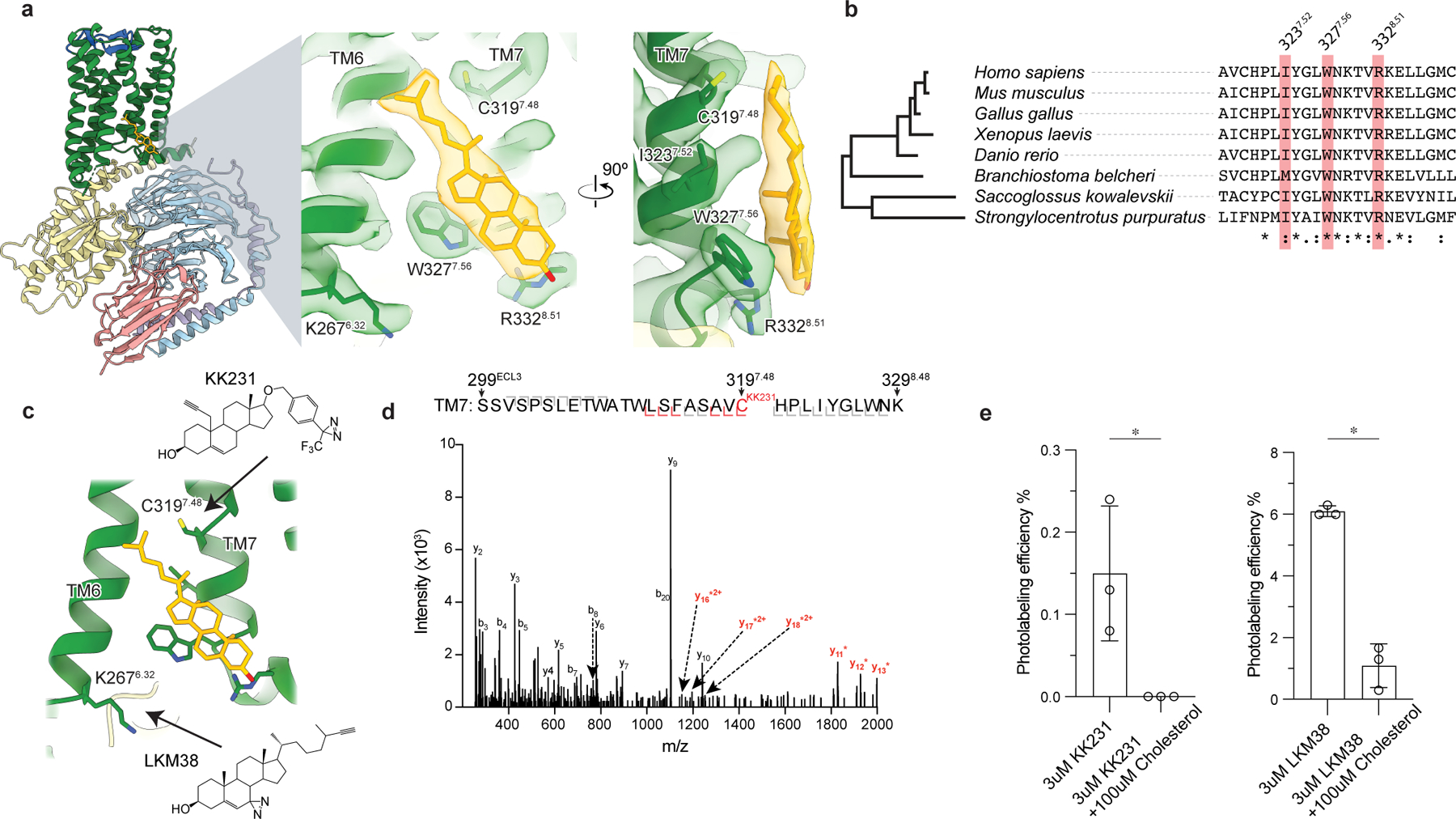
a) Close up view of cholesterol bound to the intracellular side of transmembrane helix 6 (TM6) and TM7. Three key interacting residues (I3237.52, W3277.56, R3328.51) are highlighted as sticks. b) GPR161 cholesterol binding site residues are conserved from humans to sea urchins (Strongylocentrotus purpuratus). A full alignment from these organisms used to define the dendrogram on left is shown in Extended Data Fig. 7. c) Detergent solubilized GPR161-miniGs purified without cholesterol hemisuccinate was incubated with either one of two distinct photoaffinity cholesterol analogs (KK231 or LKM38) and then crosslinked with >320 nm UV light. The resulting photo-labeled preparation was digested with trypsin and analyzed by collision-induced dissociation mass spectrometry, which revealed that KK231 labels position C3197.48 while LKM38 labels K2676.32. d) Product ion spectrum of KK231-labeled GPR161-miniGs sample with peptides mapped to TM7 and Helix 8. This peptide is modified with a mass consistent with KK231 at position C3197.48. Red brackets and peaks indicate product ions that contain the KK231 adduct. e) Photolabeling efficiency of GPR161-miniGs by KK231 and LKM238 in the absence or presence of excess unlabeled cholesterol. Data are mean ± s.d of n=3 technically independent replicates from two independently prepared protein samples (*P < 0.05, Student’s unpaired two-tailed t-test; P values: KK231 vs. KK231+cholesterol=0.0337, LKM38 vs. LKM38+cholesterol= 0.0003).
