Extended Data Fig. 2. Cryogenic electron microscopy processing of GPR161.
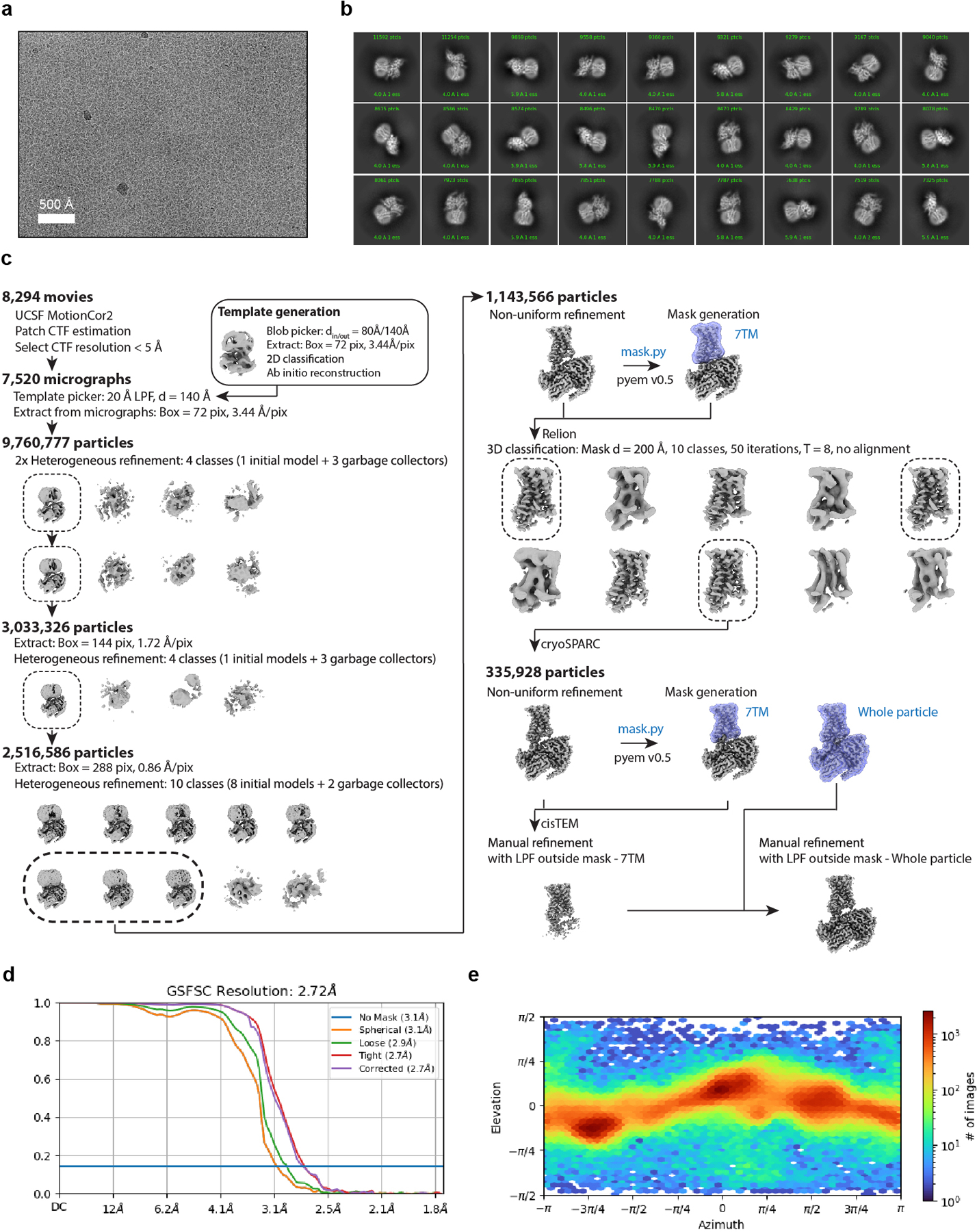
a) A representative motion-corrected cryogenic electron microscopy (cryo-EM) micrograph obtained from a Titan Krios microscope (n = 8,294). b) A subset of highly populated, reference-free 2D-class averages. c) Schematic showing the cryo-EM data processing workflow. Initial processing was performed using UCSF MotionCor2 and cryoSPARC. Particles were transferred using the pyem script package to RELION for alignment-free 3D classification. Finally, particles were processed in cisTEM using the manual refinement job type with a 7TM mask followed by a full particle mask. Dashed boxes indicated selected classes. d) Gold-standard Fourier Shell Correlation (GSFSC) curve for final refined and sharpened map computed in cryoSPARC. e) Euler angle distribution of final refined map computed in cryoSPARC.
